Potential effects of CRM1 inhibition in mantle cell lymphoma
Abstract: Mantle cell lymphoma (MCL) is an aggressive histotype of B-cell non-Hodgkin lymphoma. The disease has no known cure, which prompts the urgent need for novel therapeutic agents. Chromosomal region maintenance 1 (CRM1) may play a role in human neoplasia and serve as a novel target of cancer treatment. This study summarizes MCL pathogenesis and determines the involvement of CRM1 in the regulation of several vital signaling pathways contributing to MCL pathogenesis, including the pathways of cell cycle progression, DNA damage response, phosphoinositide kinase-3, nuclear factor-κB activation, and chromosomal stability. A preclinical study is also presented to compare the CRM1 status in MCL cell lines and primary MCL cells with normal B cells, as well as the therapeutic efficiency of CRM1 inhibition in MCL in vitro and in vivo, which make these agents potential targets of novel MCL treatments.
Key words:Chromosomal region maintenance 1 (CRM1); CRM1 inhibitor; mantle cell lymphoma
Introduction
Mantle cell lymphoma (MCL) is an aggressive disease that has been recognized as a histotype of B-cell non-Hodgkin lymphoma (NHL), a heterogeneous group of human lymphoid neoplasms with significantly increased incidence in the United States over the past three decades (1,2). Conventional chemotherapy induces MCL remission in many previously untreated patients. However, within a few years after chemotheraphy treatment, these patients experience relapse that often leads to death with a relatively short median survival duration of 5 to 7 years (3,4). Therefore, the discovery of novel therapeutic agents for MCL with low toxicity and better treatment outcomes remains a challenge.
In MCL, the non-random t[11,14][q13;32] translocation leads to cyclin D1 overexpression, which is believed to be associated with oncogenesis. However, the overexpression of cyclin D1 alone is not sufficient for MCL development, which suggests that additional genetic events are necessary for oncogenesis (5), such as the chromosomal region maintenance/exportin1/Xpo1 (CRM1) gene. CRM1 gene was first identified in the fission yeast Schizosaccharomyces pombe through genetic screening, and was determined to be involved in chromosomal structural control (6). CRM1 overexpression has been detected in several cancers (glioblastoma, ovarian, and cervical cancer) and has been associated with worse outcome (7-9).
The nucleocytoplasmic exchange of proteins (macromolecules larger than 40 kDa) is a spatially and temporally regulated process that involves several nucleocytoplasmic shuttling proteins. CRM1 is a nuclear protein export receptor belonging to the karyopherin β family of transport receptors, which transports target proteins across a guanosine triphosphate (GTP)-bound rat sarcoma (Ras)-related nuclear protein (RanGTP) gradient (10-13). CRM1 has a broad substrate range and mediates the export of leucine-rich nuclear export signal (NES)-bearing proteins through the nuclear pore complexes (NPCs) and the transfer of messenger RNAs (mRNAs) (14-18) (Figure 1).
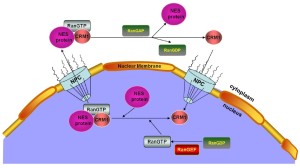
Mechanistic studies have demonstrated the importance of the CRM1 nuclear export pathway to many NES-containing signaling molecules, including P53 (19), histone deacetylase 5 (20), protein kinase 1 (21), epidermal growth factor receptor (22), and others (23,24). Given the key roles of these exported molecules in the proliferation and survival of cancer cells, including MCL cells, CRM1 could represent a new therapeutic target in MCL treatment (25-28). In this study, we summarize MCL pathogenesis and CRM1 involvement in the regulation of several vital signaling pathways contributing to MCL pathogenesis. A preclinical study is also presented to compare the CRM1 status in MCL cell lines and primary MCL cells with normal B cells, as well as the therapeutic efficiency of CRM1 inhibition in MCL in vitro and in vivo.
MCL pathogenesis
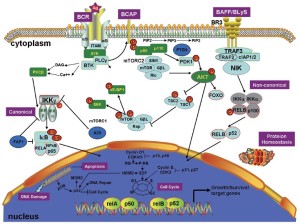
A brief overview of the relevant pathways and pathogenic mechanisms in MCL is shown in Figure 2. Cyclin D1 overexpression is the diagnostic hallmark in the majority of MCL patients (29). The aberrant B-cell receptor (BCR) (30) and B-cell activating factor signaling (31) both activate MCL cells. Furthermore, phosphoinositide kinase-3 (PI3K), Wnt, and transforming growth factor-β (TGF-β) signaling are also altered in MCL cells (). Mutations in tumor suppressors such as P53 and ataxia telangiectasia-mutated (ATM) attenuate the DNA damage response in MCL cells (33). Disordered protein homeostasis and pro-apoptotic and anti-apoptotic protein imbalances also occur in MCL. Epigenomic changes in DNA methylation and histone modifications can cause genomic instability, resulting in the aberrant expression of oncogenes and/or tumor suppressor genes, thereby contributing to MCL pathogenesis (34,35).
Regulatory roles of CRM1
Subcellular localization and function of cyclin D1
Although cyclin D1 is responsible for MCL pathogenesis, cyclin D1 overexpression does not lead to the transformation of normal lymphocytes into lymphoid malignancy in nude mice (5), whereas cyclin D1 overexpression in the nucleus induces mature B-cell lymphoma in transgenic mice (36) and drives the oncogenic transformation of murine fibroblasts in the absence of a collaborating oncogene (37,38), suggesting that the nuclear export deregulation of cyclin D1 increases its oncogenic capacity. Cyclin D1 is sequestered in the cytoplasm of mammalian cancer cells (39), where the enforced nuclear localization of cyclin D1 induces apoptosis. Thus, the subcellular localization of cyclin D1 may play a role in cell survival.
The competing processes of nuclear import and export induce cyclin D1 localization (40,41). Although the mechanisms of cyclin D1 nuclear import remain poorly characterized, the export of cyclin D1 complexes from nucleus into cytoplasm is known to be CRM1-dependent (42). Leptomycin B (LMB), a small-molecule inhibitor of CRM1, inhibits cell cycle progression and reduces cyclin D1 expression in fission yeast and mammalian cells (43). Hence, CRM1 inhibition has therapeutic potential in patients exhibiting cyclin D1 overexpression in MCL cells.
Cell cycle progression
Cell cycle dysregulation is central to MCL pathogenesis. Cyclin D1 overexpression and abnormalities in cell-cycle inhibitory genes p21WAF1, p16INK4a, and p27KIP1 have been reported in MCL (44,45). The role of CRM1 in controlling the localization and function of P21WAF1 and P27KIP1 could be exploited to treat MCL.
P21WAF1 is a cyclin-dependent kinase (CDK) inhibitor that prevents cell cycle progression at the G1 phase. The nuclear localization of the unmodified P21WAF1 is essential to elicit its anticancer functions. Two NESs are necessary to facilitate the export of P21WAF1 from the nucleus (46,47). However, the site-directed mutation of the two NESs or by LMB blocks the P21WAF1 nuclear export, which suggests the process is CRM1-mediated. P21WAF1 mainly localizes to the cytoplasm in many tumor cells (48), and cytoplasmic P21WAF1 is anti-apoptotic (49). Van der Watt et al. (9) found that CRM1 inhibition in cancer cells significantly reduces cell proliferation and increases apoptosis and P21 nuclear localization, which suggests that CRM1 is both a biomarker and a potential therapeutic target in MCL treatment.
P27KIP1, a potent cell cycle inhibitor, is significantly expressed during the G0–G1 phase transition. Sanchez-Beato et al. found that P27KIP1 expression correlated with the proliferative index in five MCL patients (50). Furthermore, low P27KIP1 expression is associated with blastoid MCL, which suggests that P27KIP1 negatively regulates the cell cycle under MCL conditions (51). The nuclear localization of P27KIP1 enables this regulatory function. However, the nuclear export of P27KIP1 is mediated by the CRM1 export receptor. Hence, CRM1 inhibition may restore the negative regulatory function of P27KIP1 in MCL cell cycle progression (52).
DNA damage-response pathways
Exogenous and endogenous stress can activate ATM, a DNA damage sensor that activates the tumor suppressor P53, which, in turn, inhibits cell cycle progression and activates DNA repair mechanisms (53). P53 is often inactivated in MCL due to its deletion or mutation (33). However, P53 activity can be regulated by its subcellular localization. P53 mislocalization arising from an aberrant import mechanism, hyperactive export, or sequestration with a cytoplasmic factor, such as the glucocorticoid receptor, has been observed in several cancers, including MCL (54,55).
One promising method of controlling cell proliferation is the relocalization of P53 to the nucleus, where it becomes active. Normally, the nuclear-cytoplasmic transportation of P53 is tightly regulated. P53 contains three nuclear localization signals (NLSs), one NES in the carboxyl terminus, and one NES in the transactivation domain (19,56-58). LMB was previously used to sequester P53 in the nucleus, leading to P53 activation and cell cycle arrest and apoptosis (59). This finding signifies the importance of modulating P53 localization and provides an impetus for developing compounds that specifically target the export of P53 from the nucleus.
PI3K pathway
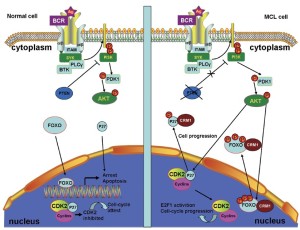
The constitutive activation of the PI3K pathway is key to MCL cell survival (29). In normal cells, phosphatase and tensin homolog (PTEN) (60), the cellular PI3K antagonist, can inhibit PI3K activation, resulting in the nuclear localization of forkhead box O (FOXO) transcription factors. In the nucleus, FOXO can activate the transcription of genes that promote cell cycle arrest and apoptosis (61). Thus, localizing FOXO to the nucleus is beneficial to controlling cell survival.
MCL cells frequently express the inactive phosphorylated form of PTEN that contributes to constitutive PI3K signaling. The constitutive activation of PI3K can also constitutively activate protein kinase B (AKT) (62). The constitutively activated AKT phosphorylates the FOXO transcription factors at multiple sites, thereby preventing FOXO-DNA binding and transcriptional activities, as well as promoting the CRM1-dependent export of FOXO from the nucleus (Figure 3) (63,64). FOXO re-localization to the nucleus, where it becomes active, is a promising method of controlling cell proliferation. Thus, CRM1 inhibition is a potential treatment for MCL.
Nuclear factor-κB activity
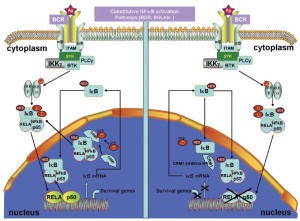
The transcriptional activator nuclear factor-κB (NF-κB) has been implicated in tumorigenesis and resistance therapy and has various roles in inflammation and immune response. The molecular mechanisms of NF-κB activation in the nucleus and the roles of NF-κB in cell proliferation and apoptosis inhibition are adequately described (65-67). In normal cells, NF-κB forms a complex with the inhibitor of κB (IκB), which masks the NLS on NF-κB and prevents NF-κB translocation to the nucleus. When IκB is phosphorylated by the IκB kinase complex and degraded by the 26S proteasome, the NLS is unmasked, and NF-κB is imported into the nucleus (60,64,68). Furthermore, the p300 acetylation of NF-κB prevents NF-κB-IκB assembly and subsequent export from the nucleus, whereas the deacetylation of NF-κB enhances its interaction with IκB and promotes subsequent export from the nucleus (69).
MCL cells express constitutively activated NF-κB. The constitutive activation of NF-κB may serve as a surrogate marker for MCL, which will be valuable in assessing the effectiveness of therapeutic agents (70). One integral component of normal NF-κB regulation is the autoregulatory negative feedback inhibition of continuous activity through the NF-κB-directed synthesis of the inhibitor of κBα (IκBα) (71). The newly synthesized IκBα enters the nucleus, removes NF-κB from the DNA, and exports NF-κB to the cytoplasm, thereby restoring the pool of inactive NF-κB-IκBα complexes (Figure 4) (72). A CRM1-dependent pathway mediates IκBα nuclear export (73). Therefore, the perturbation of the CRM1-dependent nuclear export of IκBα may attenuate constitutively activated NF-κB and cause immediate apoptosis in different cancer types (74). Therefore, the perturbation of the CRM1-dependend IκBα nuclear export induces apoptosis in MCL.
Centrosome duplication
One potential therapeutic target in MCL is centrosome aberration. Blastoid MCL is characterized by high mitotic rate of MCL cells and poor prognosis, regardless of its cytomorphological subclassification (1,75-78). It contains more chromosomal imbalances than non-blastoid MCL and exhibits high-level DNA amplifications. Moreover, blastoid MCL tends to be a tetraploid (78,79), a rare phenomenon occurring in B-cell neoplasms (80).
Chromosomal aberrations, which are common features of malignant neoplasias, can be induced by impaired DNA damage response pathways, mitotic checkpoint alterations, or centrosome aberrations. As the major microtubule-organizing centers in animal cells, centrosomes play a significant role in cell cycle progression, spindle formation, and cytokinesis (81). Neben et al. (82) found that centrosome aberrations occur more frequently in near-tetraploid MCL than in diploid MCL, suggesting that centrosome aberrations may play a role in MCL tetraploidization and are thus potential therapeutic targets in MCL, especially blastoid MCL.
In principle, centrosome aberrations could arise de novo or through the over-duplication of centrosomes within a single cell cycle, through aborted cell division, or through cell fusion (83). However, the mechanisms underlying the regulation of centrosome duplication are poorly understood. Some studies suggest that the Ran/CRM1 complex regulates nucleocytoplasmic transportation and is independently involved in mitotic spindle assembly. CRM1 may regulate the fidelity of centrosome duplication (84) by acting as a licensing factor to prevent unscheduled duplication. Hence, CRM1 inactivation either by a CRM1-specific inhibitor, such as LMB, or through the interaction of the hepatitis B virus HBx protein with its NES motif results in supernumerary centrosomes (84,85), which suggests that CRM1 may negatively regulate the initiation of centrosome duplication, possibly through its association with NES-containing proteins (Figure 5) (86,87).
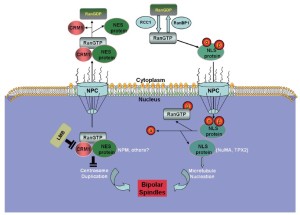
Spindle assembly
During mitosis, the supernumerary centrosomes can form multipolar spindles, which occur in many tumor types and are believed to contribute to chromosomal instability and tumorigenesis (83,88). However, some studies have shown that multipolar divisions and the resulting chromosomal instability undermine cell viability, frequently leading to cell death (89-92). Many cancer cells induce supernumerary centrosome clustering into two spindle poles, thereby enabling bipolar division, to avoid cell death. As the phenotype is specific to cancer cells, inhibiting centrosome clustering may target cancer cells selectively without affecting healthy cells.
The centrosome is composed of centrioles and pericentriolar material. A key component of the pericentriolar material and centrioles is pericentrin (PCNT2), a large conserved coiled-coil protein (93-95) that anchors γ-tubulin to the centrosomes and plays a key role in microtubule nucleation and mitotic spindle organization (93,96,97). Neben, et al. found that the expressions of four centrosome-associated proteins (PCNT2, calcium/calmodulin-dependent protein kinase II, and γ-tubulin complex-associated proteins 3 and 4) were high in MCL (98), suggesting that MCL harbors aberrant spindle assembly.
CRM1 located at centrosomes by its N-terminal CRM1, and importin beta etc. domain can interact with and regulate the localization and function of PCNT2 (99). Given the ability of PCNT2 to serve as a multifunctional scaffold for anchoring a wide range of centrosome proteins (100), it is involved in essentially all centrosome functions that center primarily around cell cycle regulation and microtubule organization. Increased PCNT2 levels can alter centrosome number, clustering, and function, thereby altering mitotic spindle organization and function, as well as causing chromosome missegregation. Centrosome clustering may enable cancer cells with multiple centrosomes to undergo relatively normal cell divisions. However, this likely leads to genetic instability, a known contributor to carcinogenesis (101-103). CRM1 inactivation can cause unscheduled centriole splitting that results in multipolar spindles. Hence, CRM1 safeguards the bipolar spindle formation by preventing unscheduled centriole splitting during mitosis (104).
Leber et al. found that the components of the chromosomal passenger complex are necessary for centrosome clustering (105). The chromosomal passenger complex is composed of aurora B and its regulatory subunits, inner centromere protein, survivin, and borealin. Survivin and aurora B are involved in the development and progression of human tumors and are thus potential targets for novel anticancer therapies (106,107). In MCL, survivin is commonly expressed in a nuclear and mitotic pattern, and its expression levels are strongly associated with tumor proliferation and patient survival (108). Dynamic nucleocytoplasmic transport regulates the subcellular localization and function of survivin and aurora B (109). Survivin and aurora B are actively excluded from the nucleus by a CRM1-mediated mechanism (110). The spatial and functional regulation of survivin by LMB abolishes the cytoprotective effect of survivin towards apoptotic executors, resulting in cell apoptosis (Figure 6) (111).
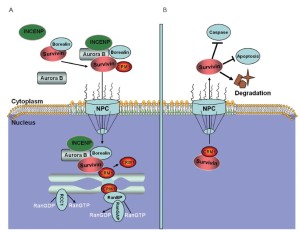
Novel CRM1 inhibitors for MCL therapy
In a previous study (112), we identified several novel CRM1 inhibitors that show potential for MCL treatments. We also found that CRM1 expression is higher in MCL cell lines and primary MCL cells than in normal B lymphocytes. Inhibiting CRM1 with small interfering RNA consequently inhibited MCL cell growth. These findings suggest that CRM1 may play a key role in the pathophysiology of MCL cells. In addition, targeting CRM1 in MCL may have therapeutic value.
Other researchers found that LMB binds covalently to cysteine 528 (Cys528) in CRM1 through a Michael-type addition reaction and abrogates the interaction between CRM1 and its cargo protein (13,113,114). Although LMB possesses strong antitumor activity in vitro, phase I trials of LMB were discontinued because of its toxicity and lack of apparent efficacy within the tolerable dose range (115). Mutka et al. (116) noted that LMB has off-target effects against proteins other than CRM1 that might contribute to its toxicity. The finding that the inhibition of CRM1 itself was not the cause of LMB toxicity is promising in terms of the development of CRM1-targeted anticancer drugs.
In order to efficiently discover novel small-molecule selective inhibitors of nuclear export (SINEs; also known as KPT-SINE compounds) that block CRM1-dependent nuclear export, Karyopharm Therapeutics applied a virtual screening workflow based on a combination of protein modeling and simulations, physicochemical filters, and high-throughput molecular docking (117,118). These CRM1-specific inhibitors are similar to the N-azolylacrylate structures developed by Daelemans et al. (119). KPT-SINE compounds are water-soluble, irreversible inhibitors of CRM1 that bind to the reactive site of the Cys528 residue. Azmi et al. (120) demonstrated that KPT-SINE compounds can induce apoptosis in resistant NHL cell lines and corresponding xenograft models. Their study verifies CRM1 as a potential therapeutic target in NHL irrespective of the functional status of P53. We found that using KPT-SINE compounds to inhibit CRM1 in MCL cell lines and primary MCL cells could significantly inhibit MCL cell growth and induce apoptosis. The oral administration of KPT-276, a KPT-SINE compound, significantly suppressed tumor growth in an MCL-bearing severe combined immunodeficient mouse model without severe toxicity (112). These findings suggest that CRM1 inhibitors are a potential novel therapy for MCL patients.
Conclusions
In conclusion, CRM1 can regulate MCL cell proliferation, cell cycle progression, DNA damage response, and chromosomal stability, making it a potential therapeutic target in MCL treatment. We found that CRM1 is overexpressed in MCL cells and that CRM1 inhibition using small interfering RNA or CRM1 inhibitors, such as KPT-SINE compounds, significantly inhibits MCL cell growth both in vitro and in vivo, making these agents potential novel treatments for MCL. Clinical studies to evaluate the therapeutic effects of KPT-SINE compounds in MCL patients are warranted.
Acknowledgements
We would like to thank Joe Munch in MD Anderson’s Department of Scientific Publications for editing the manuscript. This work was supported in part by grants from Fujian Provincial Department of Science &Technology (2009-CXB-57/ 2011J01252) and Bureau of Science & Technology of Xiamen, China (3502Z20094012).
Disclosure: The authors indicated no potential conflicts of interest.
References
- Lardelli P, Bookman MA, Sundeen J, et al. Lymphocytic lymphoma of intermediate differentiation. Morphologic and immunophenotypic spectrum and clinical correlations. Am J Surg Pathol 1990;14:752-63.
- Levine PH, Hoover R. The emerging epidemic of non-Hodgkin’s lymphoma: current knowledge regarding etiological factors. Cancer Epidemiol Biomarkers Prev 1992;1:515-7.
- Herrmann A, Hoster E, Zwingers T, et al. Improvement of overall survival in advanced stage mantle cell lymphoma. J Clin Oncol 2009;27:511-8.
- Chandran R, Gardiner SK, Simon M, et al. Survival trends in mantle cell lymphoma in the United States over 16 years 1992-2007. Leuk Lymphoma 2012;53:1488-93.
- Bodrug SE, Warner BJ, Bath ML, et al. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J 1994;13:2124-30.
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene CRM1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J Cell Biol 1989;108:1195-207.
- Noske A, Weichert W, Niesporek S, et al. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer 2008;112:1733-43.
- Shen A, Wang Y, Zhao Y, et al. Expression of CRM1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery 2009;65:153-9.
- van der Watt PJ, Maske CP, Hendricks DT, et al. The Karyopherin proteins, CRM1 and Karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int J Cancer 2009;124:1829-40.
- Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 2007;17:193-201.
- Fukuda M, Asano S, Nakamura T, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997;390:308-11.
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 1997;278:141-4.
- Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol 2012;83:1021-32.
- Kudo N, Khochbin S, Nishi K, et al. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem 1997;272:29742-51.
- Stade K, Ford CS, Guthrie C, et al. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 1997;90:1041-50.
- Yoshida M, Horinouchi S. Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann N Y Acad Sci 1999;886:23-36.
- Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 2001;294:1895-901.
- Jang BC, Munoz-Najar U, Paik JH, et al. Leptomycin B, an inhibitor of the nuclear export receptor CRM1, inhibits COX-2 expression. J Biol Chem 2003;278:2773-6.
- Stommel JM, Marchenko ND, Jimenez GS, et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 1999;18:1660-72.
- Harrison BC, Roberts CR, Hood DB, et al. The CRM1 nuclear export receptor controls pathological cardiac gene expression. Mol Cell Biol 2004;24:10636-49.
- Saji M, Vasko V, Kada F, et al. Akt1 contains a functional leucine-rich nuclear export sequence. Biochem Biophys Res Commun 2005;332:167-73.
- Lo HW, Ali-Seyed M, Wu Y, et al. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem 2006;98:1570-83.
- Ramaswami S, Manna S, Juvekar A, et al. Chromatin immunoprecipitation analysis of NFkappaB transcriptional regulation by nuclear IkappaBalpha in human macrophages. Methods Mol Biol 2012;809:121-34.
- Gustafsson MO, Hussain A, Mohammad DK, et al. Regulation of nucleocytoplasmic shuttling of bruton’s tyrosine kinase (Btk) through a novel SH3-dependent interaction with ankyrin repeat domain 54 (ANKRD54). Mol Cell Biol 2012;32:2440-53.
- Gray LJ, Bjelogrlic P, Appleyard VC, et al. Selective induction of apoptosis by leptomycin B in keratinocytes expressing HPV oncogenes. Int J Cancer 2007;120:2317-24.
- Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res 2009;15:752-7.
- Dey A, Tergaonkar V, Lane DP. Double-edged swords as cancer therapeutics: simultaneously targeting p53 and NF-kappaB pathways. Nat Rev Drug Discov 2008;7:1031-40.
- Ranganathan P, Yu X, Na C, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 2012;120:1765-73.
- Pérez-Galán P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood 2011;117:26-38.
- Pighi C, Gu TL, Dalai I, et al. Phospho-proteomic analysis of mantle cell lymphoma cells suggests a pro-survival role of B-cell receptor signaling. Cell Oncol (Dordr) 2011;34:141-53.
- Fu L, Lin-Lee YC, Pham LV, et al. BAFF-R promotes cell proliferation and survival through interaction with IKKbeta and NF-kappaB/c-Rel in the nucleus of normal and neoplastic B-lymphoid cells. Blood 2009;113:4627-36.
- Rizzatti EG, Falcao RP, Panepucci RA, et al. Gene expression profiling of mantle cell lymphoma cells reveals aberrant expression of genes from the PI3K-AKT, WNT and TGFbeta signalling pathways. Br J Haematol 2005;130:516-26.
- Greiner TC, Moynihan MJ, Chan WC, et al. p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood 1996;87:4302-10.
- Leshchenko VV, Kuo PY, Shaknovich R, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood 2010;116:1025-34.
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27-36.
- Gladden AB, Woolery R, Aggarwal P, et al. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene 2006;25:998-1007.
- Alt JR, Cleveland JL, Hannink M, et al. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev 2000;14:3102-14.
- Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res 2003;63:7056-61.
- Alao JP, Gamble SC, Stavropoulou AV, et al. The cyclin D1 proto-oncogene is sequestered in the cytoplasm of mammalian cancer cell lines. Mol Cancer 2006;5:7.
- Alt JR, Gladden AB, Diehl JA. p21(Cip1) Promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J Biol Chem 2002;277:8517-23.
- Lin X, Nelson P, Gelman IH. SSeCKS, a major protein kinase C substrate with tumor suppressor activity, regulates G(1)-->S progression by controlling the expression and cellular compartmentalization of cyclin D. Mol Cell Biol 2000;20:7259-72.
- Benzeno S, Diehl JA. C-terminal sequences direct cyclin D1-CRM1 binding. J Biol Chem 2004;279:56061-6.
- Tsuchiya A, Tashiro E, Yoshida M, et al. Involvement of protein phosphatase 2A nuclear accumulation and subsequent inactivation of activator protein-1 in leptomycin B-inhibited cyclin D1 expression. Oncogene 2007;26:1522-32.
- Pinyol M, Hernandez L, Cazorla M, et al. Deletions and loss of expression of p16INK4a and p21Waf1 genes are associated with aggressive variants of mantle cell lymphomas. Blood 1997;89:272-80.
- Letestu R, Ugo V, Valensi F, et al. Prognostic impact of p27KIP1 expression in cyclin D1 positive lymphoproliferative disorders. Leukemia 2004;18:953-61.
- Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 2000;256:213-24.
- Hwang CY, Kim IY, Kwon KS. Cytoplasmic localization and ubiquitination of p21(Cip1) by reactive oxygen species. Biochem Biophys Res Commun 2007;358:219-25.
- Zhou BP, Liao Y, Xia W, et al. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol 2001;3:245-52.
- Asada M, Yamada T, Ichijo H, et al. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J 1999;18:1223-34.
- Sánchez-Beato M, Saez AI, Martinez-Montero JC, et al. Cyclin-dependent kinase inhibitor p27KIP1 in lymphoid tissue: p27KIP1 expression is inversely proportional to the proliferative index. Am J Pathol 1997;151:151-60.
- Izban KF, Alkan S, Singleton TP, et al. Multiparameter immunohistochemical analysis of the cell cycle proteins cyclin D1, Ki-67, p21WAF1, p27KIP1, and p53 in mantle cell lymphoma. Arch Pathol Lab Med 2000;124:1457-62.
- Susaki E, Nakayama K, Nakayama KI. Cyclin D2 translocates p27 out of the nucleus and promotes its degradation at the G0-G1 transition. Mol Cell Biol 2007;27:4626-40.
- Lavin MF. ATM: the product of the gene mutated in ataxia-telangiectasia. Int J Biochem Cell Biol 1999;31:735-40.
- Kim IS, Kim DH, Han SM, et al. Truncated form of importin alpha identified in breast cancer cell inhibits nuclear import of p53. J Biol Chem 2000;275:23139-45.
- Sengupta S, Vonesch JL, Waltzinger C, et al. Negative cross-talk between p53 and the glucocorticoid receptor and its role in neuroblastoma cells. EMBO J 2000;19:6051-64.
- Shaulsky G, Goldfinger N, Ben-Ze’ev A, et al. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol 1990;10:6565-77.
- Liang SH, Hong D, Clarke MF. Cooperation of a single lysine mutation and a C-terminal domain in the cytoplasmic sequestration of the p53 protein. J Biol Chem 1998;273:19817-21.
- Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 2001;292:1910-5.
- Laín S, Midgley C, Sparks A, et al. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp Cell Res 1999;248:457-72.
- Beg AA, Ruben SM, Scheinman RI, et al. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev 1992;6:1899-913.
- Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science 1996;274:782-4.
- Ferrigno P, Silver PA. Regulated nuclear localization of stress-responsive factors: how the nuclear trafficking of protein kinases and transcription factors contributes to cell survival. Oncogene 1999;18:6129-34.
- Van Antwerp DJ, Martin SJ, Kafri T, et al. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science 1996;274:787-9.
- Henkel T, Zabel U, van Zee K, et al. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-kappa B subunit. Cell 1992;68:1121-33.
- Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 1999;18:6938-47.
- Perkins ND. The Rel/NF-kappa B family: friend and foe. Trends Biochem Sci 2000;25:434-40.
- Karin M, Cao Y, Greten FR, et al. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2002;2:301-10.
- Ganchi PA, Sun SC, Greene WC, et al. I kappa B/MAD-3 masks the nuclear localization signal of NF-kappa B p65 and requires the transactivation domain to inhibit NF-kappa B p65 DNA binding. Mol Biol Cell 1992;3:1339-52.
- Chen LF, Greene WC. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J Mol Med (Berl) 2003;81:549-57.
- Pham LV, Tamayo AT, Yoshimura LC, et al. Inhibition of constitutive NF-kappa B activation in mantle cell lymphoma B cells leads to induction of cell cycle arrest and apoptosis. J Immunol 2003;171:88-95.
- Chiao PJ, Miyamoto S, Verma IM. Autoregulation of I kappa B alpha activity. Proc Natl Acad Sci USA 1994;91:28-32.
- Arenzana-Seisdedos F, Turpin P, Rodriguez M, et al. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J Cell Sci 1997;110:369-78.
- Huang TT, Kudo N, Yoshida M, et al. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc Natl Acad Sci U S A 2000;97:1014-9.
- O’Connor S, Shumway S, Miyamoto S. Inhibition of IkappaBalpha nuclear export as an approach to abrogate nuclear factor-kappaB-dependent cancer cell survival. Mol Cancer Res 2005;3:42-9.
- Jaffe ES, Bookman MA, Longo DL. Lymphocytic lymphoma of intermediate differentiation--mantle zone lymphoma: a distinct subtype of B-cell lymphoma. Hum Pathol 1987;18:877-80.
- Fisher RI, Dahlberg S, Nathwani BN, et al. A clinical analysis of two indolent lymphoma entities: mantle cell lymphoma and marginal zone lymphoma (including the mucosa-associated lymphoid tissue and monocytoid B-cell subcategories): a Southwest Oncology Group study. Blood 1995;85:1075-82.
- Zukerberg LR, Medeiros LJ, Ferry JA, et al. Diffuse low-grade B-cell lymphomas. Four clinically distinct subtypes defined by a combination of morphologic and immunophenotypic features. Am J Clin Pathol 1993;100:373-85.
- Ott MM, Ott G, Kuse R, et al. The anaplastic variant of centrocytic lymphoma is marked by frequent rearrangements of the bcl-1 gene and high proliferation indices. Histopathology 1994;24:329-34.
- Beà S, Ribas M, Hernandez JM, et al. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood 1999;93:4365-74.
- Ott G, Kalla J, Ott MM, et al. Blastoid variants of mantle cell lymphoma: frequent bcl-1 rearrangements at the major translocation cluster region and tetraploid chromosome clones. Blood 1997;89:1421-9.
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol 2005;21:411-34.
- Krämer A, Schweizer S, Neben K, et al. Centrosome aberrations as a possible mechanism for chromosomal instability in non-Hodgkin’s lymphoma. Leukemia 2003;17:2207-13.
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer 2002;2:815-25.
- Forgues M, Difilippantonio MJ, Linke SP, et al. Involvement of Crm1 in hepatitis B virus X protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol Cell Biol 2003;23:5282-92.
- Forgues M, Marrogi AJ, Spillare EA, et al. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J Biol Chem 2001;276:22797-803.
- Fukasawa K, Choi T, Kuriyama R, et al. Abnormal centrosome amplification in the absence of p53. Science 1996;271:1744-7.
- Fodde R, Kuipers J, Rosenberg C, et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol 2001;3:433-8.
- Krämer A, Neben K, Ho AD. Centrosome replication, genomic instability and cancer. Leukemia 2002;16:767-75.
- Weaver BA, Silk AD, Montagna C, et al. Aneuploidy: acts both oncogenically and as a tumor suppressor. Cancer Cell 2007;11:25-36.
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009;460:278-82.
- Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol 2001;11:18-21.
- Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA 2004;101:8699-704.
- Doxsey SJ, Stein P, Evans L, et al. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 1994;76:639-50.
- Jurczyk A, Gromley A, Redick S, et al. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol 2004;166:637-43.
- Miyoshi K, Asanuma M, Miyazaki I, et al. Characterization of pericentrin isoforms in vivo. Biochem Biophys Res Commun 2006;351:745-9.
- Purohit A, Tynan SH, Vallee R, et al. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J Cell Biol 1999;147:481-92.
- Zimmerman WC, Sillibourne J, Rosa J, et al. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell 2004;15:3642-57.
- Neben K, Ott G, Schweizer S, et al. Expression of centrosome-associated gene products is linked to tetraploidization in mantle cell lymphoma. Int J Cancer 2007;120:1669-77.
- Liu Q, Yu J, Zhuo X, et al. Pericentrin contains five NESs and an NLS essential for its nucleocytoplasmic trafficking during the cell cycle. Cell Res 2010;20:948-62.
- Delaval B, Doxsey SJ. Pericentrin in cellular function and disease. J Cell Biol 2010;188:181-90.
- Quintyne NJ, Reing JE, Hoffelder DR, et al. Spindle multipolarity is prevented by centrosomal clustering. Science 2005;307:127-9.
- Rebacz B, Larsen TO, Clausen MH, et al. Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen. Cancer Res 2007;67:6342-50.
- Kwon M, Godinho SA, Chandhok NS, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 2008;22:2189-203.
- Budhu AS, Wang XW. Loading and unloading: orchestrating centrosome duplication and spindle assembly by Ran/Crm1. Cell Cycle 2005;4:1510-4.
- Leber B, Maier B, Fuchs F, et al. Proteins required for centrosome clustering in cancer cells. Sci Transl Med 2010;2:33ra38
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer 2003;3:46-54.
- Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol 2005;15:241-50.
- Martinez A, Bellosillo B, Bosch F, et al. Nuclear survivin expression in mantle cell lymphoma is associated with cell proliferation and survival. Am J Pathol 2004;164:501-10.
- Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res 2003;282:59-69.
- Rodriguez JA, Lens SM, Span SW, et al. Subcellular localization and nucleocytoplasmic transport of the chromosomal passenger proteins before nuclear envelope breakdown. Oncogene 2006;25:4867-79.
- Chan KS, Wong CH, Huang YF, et al. Survivin withdrawal by nuclear export failure as a physiological switch to commit cells to apoptosis. Cell Death Dis 2010;1:e57.
- Zhang K, Pham LV, Zhang L, et al. Novel CRM-1 Inhibitors for Therapy In Mantle Cell Lymphoma. ASH Annual Meeting Abstracts 2011;118:2734.
- Yashiroda Y, Yoshida M. Nucleo-cytoplasmic transport of proteins as a target for therapeutic drugs. Curr Med Chem 2003;10:741-8.
- Kudo N, Matsumori N, Taoka H, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 1999;96:9112-7.
- Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer 1996;74:648-9.
- Mutka SC, Yang WQ, Dong SD, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res 2009;69:510-7.
- Draetta GF, Shacham S, Kauffman M, et al. Cytotoxicity of novel, small molecule, CRM1-selective inhibitors of nuclear export (SINE) in colorectal cancer (CRC) cells. ASCO Meeting Abstracts 2011;29:e14091.
- Shacham S, Turner JG, Nir R, et al. Preclinical Development of Small-Molecule CRM1 Inhibitors as Novel Therapy for the Treatment of Myeloma and Other Hematological Malignancies. ASH Annual Meeting Abstracts 2010;116:3012.
- Daelemans D, Afonina E, Nilsson J, et al. A synthetic HIV-1 Rev inhibitor interfering with the CRM1-mediated nuclear export. Proc Natl Acad Sci U S A 2002;99:14440-5.
- Azmi AS, Al-Katib A, Aboukameel A, et al. Development of a Novel Small Molecule CRM-1 Inhibitor for Non Hodgkin’s Lymphoma. ASH Annual Meeting Abstracts 2011;118:598.


