Analysis of miR-205 and miR-155 expression in the blood of breast cancer patients
Introduction
MicroRNAs (miRNAs) are small (19-25 ribonucleotides), single-stranded non-coding RNAs found in plant, invertebrate and vertebrate genomes (1). They regulate gene expression at the post-transcriptional level by inducing mRNA degradation or translational inhibition of specific messenger RNA (mRNA) targets (2,3). miRNAs are involved in regulating cell differentiation, cell cycle progression and apoptosis. More than 900 miRNAs have been identified in humans to date, and each miRNA is assigned a numerical identifier (4).
Deregulation of miRNA expression has been demonstrated in many types of cancers (5-7). Many miRNAs are located at fragile sites or cancer-associated regions of the genome, which may explain why there is a correlation between tumorigenesis and aberrant expression of certain miRNAs (8). Although a large number of miRNAs have been identified to date, the role for many of them in tumorigenesis and their underlying mechanisms remain to be determined. miR-205, located in 1q32.2, was shown to be overexpressed in head and neck cancer compared with lung, breast, colorectal, prostate, and pancreatic cancer cell lines (9). Yanaihara et al. (10) have shown that miRNA expression profiles are diagnostic and prognostic markers for lung cancer. Upregulation of miR-205 was observed in bladder cancer (11) as well as in adult mouse corneal and footpad epithelium (12).
Taylor and Gercel-Taylor (13) found that miRNA from both ovarian tumor cells and exosomes from the same patients were positive for 218 of 467 mature microRNAs analyzed. miRNA profiling of circulating tumor exosomes could thus potentially be used as a surrogate diagnostic marker for biopsy profiling, which could allow its utility to be extended to screening asymptomatic populations. Chen et al. (14) sequenced all serum miRNAs of healthy Chinese subjects and found over 100 and 91 serum miRNAs in male and female subjects, respectively. They also identified specific expression patterns of serum miRNAs from lung cancer, colorectal cancer, and diabetes, providing evidence that serum miRNAs contain fingerprints of various diseases. The expression of miR-205 in the serum of breast cancer has not been reported. In this study, we report that miR-205 is downregulated in breast tumor tissues, which is in good agreement with a previous report (15). Importantly, ectopic expression of miR-205 significantly enhance apoptosis and anchorage-independent growth of MCF-7 breast cancer cells. We further demonstrated that miR-205 is down-regulated and miR-155 is up-regulated in breast cancer serum. This observation may be a new biomarker for breast cancer diagnosis and screening.
Materials and methods
Main reagents
Penicillin-streptomycin solution, ficoll-hypaque solution, trypsin-EDTA solution, RPMI-1640 medium, Dulbecco’s modified Eagle’s medium (DMEM/F-12), and 1% antibiotic-antimycotic solution were obtained from Life Technologies GIBCO, Grand Island, NY, USA. LiporeetamineTMZ000 was purchased from Invitrogen. OPTI-MEM was obtained from GIBCO. AnllexinV-FITC apoptosis detection kit was obtained from KeyGEN BioTECH, Nanjin, China. hsa-miR-205 mimics and cy3 dye-labeled miR negative control was purchased from RiboBio Co. Ltd., Guangzhou, Guangdong. 10% fetal bovine serum was obtained from HyClone, Logan, UT.
Samples and cases
For the miRNA microarray analysis, fresh samples from four cases of human BC and paired normal adjacent tissues (NATs, >2 cm from cancer tissue) were obtained from Tianjin cancer hospital between 2007 and 2009. The patients aged 50-55, two samples with ER(+)PR(+)Her-2(+) were collected and the others with ER(-)PR(-)Her-2(-).The fresh specimens were stored at 4 °C for 24 h in RNA Later (Ambion Inc.) and then at −80 °C liquid nitrogen until further use. The samples used were not subjected to preoperative radiotherapy and/or chemotherapy and were diagnosed as infiltrating carcinoma by pathology. Before RNA extraction, sections were stained with H&E for histological diagnosis and tumor cell evaluation. Only those cases with >70% tumor cell population in the section were used in this study. Two independent sets of plasma samples were used. Plasma samples from age- and gender-matched healthy donors and breast cancer patients (Stage I-IV) were obtained at the Tianjin Medical University Cancer Institute and Hospital, Tianjin, China, between 2007 and 2009. Twenty of these individuals were breast cancer patients, and 10 were normal healthy individuals with no evidence of any disease. All blood samples were collected prior to any therapeutic procedures, including surgery. The plasma was isolated within an hour by centrifugation at 1,500× g at room temperature for 20 minutes and stored as multiple aliquots in fresh tubes at −80 °C.
MiRNA microarray experiments
For miRNA microarray analysis, total RNA from fresh BC tissue was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA samples were analyzed by CapitalBio (CapitalBio Corp.). Procedures were performed as described in detail on the CapitalBio website (
RNA isolation and quantitative RT-PCR in plasma
Total RNA containing small RNA was isolated from plasma using the Trizol LS reagent (Invitrogen Life Technologies) according to the manufacturer’s protocol with the following modifications. The Trizol LS reagent was mixed with plasma in a 3:1 ratio and incubated for 5 minutes. After the addition of chloroform, tubes were shaken well and centrifuged to separate the upper aqueous phase, which was carefully transferred to a fresh tube. Isopropanol was then added to the aqueous phase for 30 minutes followed by centrifugation at 12,000× g for 10 minutes. The RNA precipitate was washed with 75% ethanol and centrifuged at 7,500× g for 5 minutes. miRNA-specific stem-loop primers (Table 1) and other primers were obtained from RiboBio Co. Ltd, Guangzhou, Guangdong. Subsequently, quantitative real-time polymerase chain reaction (qRT-PCR) was performed with an ABI Prism 7000 sequence detection system (Applied Biosystems). RT PCR system: (1×) 10 mM dNTPs (0.15 L), Multi-Scribe Reverse Transcriptase (1 L), 10× RT buffer (1.5 L), RNase Inhibitor (0.19 L), nuclease-free water (6.66 L), RNA sample (2.5 L), 5× RT primer (3 L); 16 °C for 30 min, 42 °C for 30 min, 85 °C for 5 min, 4 °C forever. Pre-amplification system: (1×) TaqMan PreAmp Master Mix (2×) (10 L), nuclease-free (7.67 L) water, 20× primer (Applied Biosystem 1:50 dillution) (1 L), miRNA RT product (1.33 L); Hold 95 °C 10 min, Hold 55 °C 2 min, Hold 72 °C 2 min, 95 °C 15 sec, 60 °C 4 min, Repeat step 4 and 5 for 12 cycles, Hold 99.0 °C 10 min. Real time PCR system: 2× TaqMan Universal PCR Master Mix (10 µL), 20× PCR Primer (1 µL), nuclease-free water (7.67 µL) and miRNA RT product (1:50 dilution) (1.33 L). qRT-PCR was then performed at 95 °C for 10 minutes, 95 °C for 15 seconds and 60 °C for 60 seconds, with the last two steps repeated for a total of 45 cycles. Each reaction was performed in triplicate. miRNA expression was defined based on the threshold cycle (Ct), and relative expression levels were calculated as 2-[(Ct of miR-205) – (Ct of U6)] after normalization with reference to expression of U6.

Full table
Cell culture
The MCF-7 human breast cancer cell line was obtained from the center laboratory of Tianjin cancer hospital and was maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic solution. Cells were grown to confluence at 37 °C in 5% CO2 atmosphere.
Synthesis and transfection of miRNA mimics
The miR-205 mimics and negative control were purchased from RiboBio (RiboBio Co. Ltd, Guangzhou, Guangdong). Transfection of microRNA mimics and the negative control were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. The third group was treated with only lipo-2000, we define it as blank group.
Apoptosis assay
12, 24, 48, and 72 hours after transfection, 40 µL of MTT (5 mg/mL) was added to each well of a 96-well plate and the cells were cultured for 4 h. The MTT medium mixture was then discarded and 100 µL of dimethyl sulfoxide (DMSO) was added to each well. Absorbance was measured at 490 nm using a multi-well spectrophotometer (Bio-Tek, USA). Each reaction was performed in triplicate. For cell cycle analysis, the cells were resuspended in PBS and then fixed in ethanol at −20 °C overnight. The cells were washed with PBS and resuspended in Staining Solution (50 µg/mL of propidium iodide, 1 mg/mL of RNase A, 0.1% Triton X-100 in PBS). The stained cells (1×106) were then analyzed with a flow cytometer. Cell apoptosis was detected using an AnllexinV-FITC apoptosis detection kit (KeyGEN BioTECH, Nanjin, China) according to the manufacturer’s protocol and FACS analysis.
Statistical analysis
Statistical comparisons were performed with SAM software (SAM version 2.1,
The relative miRNA levels were quantified using 2-△△T; the other data are presented as 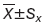 from at least three separate experiments. Multiple group comparisons were performed with ANOVA (P<0.05).
from at least three separate experiments. Multiple group comparisons were performed with ANOVA (P<0.05).
Results
MiRNA expression profile of BC
SAM analysis generated a list of differentially expressed miRNAs at FDR=0 (0% false discovery rate). We identified one miRNA that was up-regulated greater than two-fold (miR-155) in BC compared with NAT. Nine miRNAs were down-regulated greater than two-fold: let-7b, miR-381, miR-10b, miR-125a-5p, miR-335, miR-205 and miR-145 (Table 2).
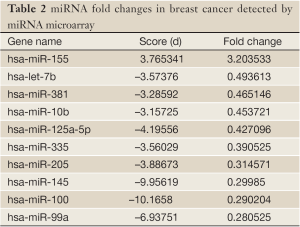
Full table
MiR-205 and miR-155 expression in the serum of BC patients versus control
Total RNA was isolated from each sample and miRNA content was determined by qRT-PCR. We profiled miRNA expression in the serum of 20 BC patients and 10 healthy controls. miR-205 expression on average was down-regulated in BC patients compared with the matched healthy controls (Figure 1; P<0.05) while miR-155 expression was up-regulated (Figure 1; P<0.05). We investigated associations between miR-205 and the clinicopathologic features of the BC patients but found no significant correlation.
miR-155 expression had significant difference in the clinical stage, molecular type, Ki-67 and p53 status of BC patients (P<0.05) but no significant difference in age, and histological grade. The relative expression of miR-155 was strongest in phase III, followed by phase II and phase I; it was strongest in “triple negative” subtype, followed by HER-2(+), luminal B, and luminal A; and finally, it was stronger in Ki-67(+) than in Ki-67(-) and stronger in p53(-) than in p53(+) patients (Table 3).
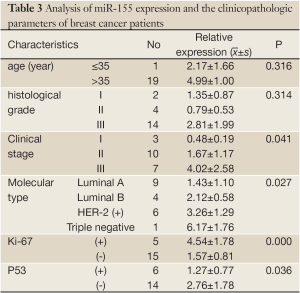
Full table
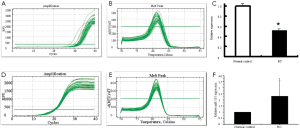
Growth inhibition of MCF-7 cells by miR-205
The cellular activity in each group was determined using the MTT method 12, 24, 48, and 72 hours after transfection, from which growth curves were established and the relative cellular proliferation activities were calculated (Figure 2). It was found that the relative proliferation activity was 2.28±0.25%, 4.35±0.28%, and 4.62±0.31% in the 72-hour transfection group, negative control group, and blank group, respectively. Compared with the negative control group and blank group, the growth of MCF-7 cells was significantly inhibited in the transfection groups (P<0.01); however, no such difference was found between the negative control group and the blank group (P>0.05).
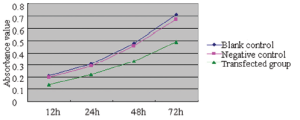
MiR-205 induces apoptosis of MCF-7 cells
We conducted an MCF-7 cell proliferation assay to determine whether miR-205 suppresses MCF-7 cell entry into S phase. A comparison of treatments between the three groups (Figure 3) indicated that the G0/G1 ratio was increased and S phase was lower, but there was no significant difference between the groups (P>0.05). Moreover, we examined cell apoptosis in MCF-7 cells using an AnllexinV-FITC apoptosis detection kit and found that levels of cell apoptosis were increased in cells transfected with miR-205 mimics versus other groups (Figure 4). Taken together, our data indicate that miR-205 suppresses MCF-7 cell growth via enhancement of apoptosis.
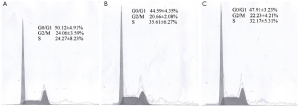

Discussion
miRNAs were first linked with cancer in 2002 initially in chronic lymphocytic leukemia (CLL) and subsequently in many other types of cancer, including BC (19,20). miR-155 was first found as an oncogene and was highly expressed in many types of tumors (21). BC, the second-leading cause of cancer-related deaths in women, is expected to account for 26% of new cancer diagnoses in 2008 (22). Several miRNAs are associated with BC. Patients with low miR-335 and miR-126 have been observed to have shorter median times to metastatic relapse (23). Iorio et al. (24) used a miRNA microarray to evaluate miRNA expression profiles of 10 normal and 76 neoplastic breast tissues. They found that miR-10b, miR125b and miR-145 were down-regulated in breast cancer while miR-21 and miR-155 were up-regulated. Based on microarray expression experiments, we found that miR-155 was highly expressed in BC while miR-145, miR-335, miR-10b, miR-125a and miR-205 were down-regulated. These results were similar to another study (24). miR-205 was found to be higher in ER+PR+Her2+ BC relative to other subtypes (25,26); however, other studies observed high miR-205 expression in ER−PR−Her2− tumors, but low expression in other tumor types, and low miR-205 expression in metastatic BC cell lines (5). Likewise, other studies found low miR-205 expression in clinical samples of metastatic BC compared to non-metastatic cancers (24). In our study, however, there are only four paired tissues; thus, we cannot analyze specific subsets.
Although tumor tissue miRNA have been demonstrated to be associated with the development and prognosis of tumors in many studies, the complex maneuver and large trauma to patients during its determination make it difficult to be applied to clinical practice. On the contrary, the levels of tumor associated microRNAs in the serum of the peripheral blood are much easier to determine, and therefore are more feasible for clinical application. Many articles have reported the association between serum miRNA and tumors, which provides a solid foundation for the application of serum miRNA as a molecular marker in the early diagnosis and prognosis of tumors (13,14). miRNAs are present in human plasma in a remarkably stable form that is protected from endogenous RNase activity (27,28). Plasma miR-92 was used to diagnosis the colorectal cancer (CRC), with a sensitivity of 89% and specificity of 70% in discriminating CRC from control subjects. It can be a potential non-invasive molecular marker for CRC screening (29). Based on the differentially expressed miRNA obtained by chip screening, we further determined the expression of miR-205 and miR-155 in the serum of 20 patients with BC and in control samples using real time technology. It was found that these two miRNAs were differentially expressed within the serum of BC patients; furthermore, miR-155 was associated with the clinical stages of BC and with ki-67, p53 and molecular type. Its relative expression in serum was highest in triple negative BC, followed by HER-2(+), luminal B, and luminal A. In a multi-factor correlation analysis among 49 BC patients who had received adriamycin monotherapy, Sorlie et al. found that the survival rates significantly differed among patients with different molecular types: luminal A > luminal B > triple-negative and HER-2+/ER-group (30). Therefore, serum miR-155 levels are correlated with malignancy and survival rates of BC. Ki-67 is a nuclear antigen expressed in proliferative cells. Ki-67 expression represents the proliferative activity of tumor cells and therefore is often used as a reliable marker of proliferation. BC positive for Ki-67 tends to be more malignant. In such cases, tumor cells had high proliferative activity, strong invasion potential, and a high probability for metastasis; therefore, such patients usually have poor prognosis. Patients with high Ki-67 label index had significantly decreased disease-free survival and overall survival (31). Wild-type p53 gene is a key tumor suppressor gene. It can trigger cell cycle arrest, induce apoptosis, and promote differentiation (32). After genetic mutation, p53 will become mutant p53, which can induce the inhibition of cell apoptosis, uncontrolled cellular proliferation and ultimately cause the cancer development. In our current study, serum miR-155 significantly differed in terms of Ki-67 and p53 (P<0.01), with the relative expression of Ki-67(+) larger than Ki-67(-) and that that of P53 (-) larger than P53 (+), indicating that serum miR-155 levels were associated with tumor cell proliferation, invasion, apoptosis and prognosis. High miR-155 expression was detected when tumor cells had high proliferative activity and decreased apoptotic capability.
Studies have shown that many miRNAs can promote or inhibit tumor cell growth (33). For example, transfection of anti-miR-199a oligonucleotides into cervical cancer cells suppressed cell growth in vitro (34), and inhibition of miR-141 decreased cell growth in human cholangiocarcinoma cell lines (35). Our miRNA profiling suggests that miR-205 is specifically down-regulated in BC. Moreover, miR-205 expression is also lower in BC serum compared to healthy controls, suggesting that miR-205 is a potential tumor suppressor in BC. In support of this notion, we demonstrate that ectopic expression of miR-205 suppresses proliferation and increases breast tumor cell apoptosis, similar to the result of Wu (36). One study found that inhibition of MCF-7 cell growth was related to miR-205 loss in fragile sites (8).
The efficient extraction and accurate identification of miRNAs from serum represents a key first step toward the development of a noninvasive blood-based detection test for BC. Screening serum for miRNAs that predict the presence of breast cancer is feasible and may be useful for BC detection. Furthermore, it will provide useful information for evaluating BC staging and prognosis.
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 30872955).
Disclosure: The authors declare no conflict of interest.
References
- Wiwanitkit S, Wiwanitkit V. MicroRNA from tuberculosis RNA: A bioinformatics study. J Thorac Dis 2012;4:296-7.
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31.
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet 2001;2:110-9.
- Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008;36:D154-8.
- Chen J, Wang B. The roles of miRNA-143 in colon cancer and therapeutic implications. Transl Gastrointest Cancer 2012;1:169-74.
- Porkka KP, Pfeiffer MJ, Waltering KK, et al. MicroRNA expression profiling in prostate cancer. Cancer Res 2007;67:6130-5.
- Meng F, Francis H, Alpini G. Non-coding RNAs in human liver malignancies, critical regulators for cancer stemness? Transl Gastrointest Cancer 2012;1:1-4.
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004.
- Jiang J, Lee EJ, Gusev Y, et al. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 2005;33:5394-403.
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98.
- Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol 2007;25:387-92.
- Ryan DG, Oliveira-Fernandes M, Lavker RM, et al. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis 2006;12:1175-84.
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21.
- Chen X, Ba Y, Ma L, Cai X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006.
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70.
- Thomson JM, Parker J, Perou CM, et al. A custom microarray platform for analysis of microRNA gene expression. Nat Methods 2004;1:47-53.
- Castoldi M, Schmidt S, Benes V, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA 2006;12:913-20.
- He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A 2005;102:19075-80.
- Mirnezami AH, Pickard K, Zhang L, et al. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. Eur J Surg Oncol 2009;35:339-47.
- Yang L, Belaguli N, Berger DH. MicroRNA and colorectal cancer. World J Surg 2009;33:638-46.
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61.
- Wedam SB, Yang SX. Neoadjuvant bevacizumab and chemotherapyin breast cancer. Transl Cancer Res 2012;1:57-8.
- Tavazoie SF, Alarcón C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008;451:147-52.
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70.
- Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 2006;5:24.
- Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 2007;67:11612-20.
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8.
- Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 2009;106:4402-7.
- Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009;58:1375-81.
- Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869-74.
- Mao XY, Fan CF, Zheng HC, et al. p53 nuclear accumulation and ERalpha expression in ductal hyperplasia of breast in a cohort of 215 Chinese women. J Exp Clin Cancer Res 2010;29:112.
- Van Dyke T. p53 and tumor suppression. N Engl J Med 2007;356:79-81.
- Schumacher U, Adam E, Feldhaus S, et al. Cell differentiation and chemotherapy influence p53 and Mdm2 immunoreactivity in human HT29 colon cancer cells grown in scid mice. Cancer Lett 2001;166:215-21.
- Lee JW, Choi CH, Choi JJ, et al. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res 2008;14:2535-42.
- Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006;130:2113-29.
- Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 2009;19:439-48.


