PIK3CA mutation in Chinese patients with lung squamous cell carcinoma
Introduction
Lung cancer is the most frequent cause of cancer-related death worldwide. Despite therapeutic advances, the overall 5-year survival remains only 15% (1). Novel treatment strategies are, therefore, needed. One promising treatment strategy involves the further subdivision of non-small cell lung cancer (NSCLC) into clinically relevant molecular subsets, according to a classification schema based on specific so-called driver mutations to perform molecular targeted therapies (2,3). For instance, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (gefitinib and erlotinib) and anaplastic lymphoma kinase (ALK) inhibitor (crizotinib) have dramatically improved the response and progression-free survival of NSCLC patients harboring EGFR sensitive mutations and echinoderm microtubule-associated protein like-4 (EML4)-ALK fusion gene (4-6). These mutations occur in genes that encode signaling proteins crucial for cellular proliferation and survival. Mutant oncogenes drive tumor formation and maintenance (7,8). Cancers might rely on the expression of these single-mutant oncogenes for survival. This concept is also called oncogene addiction (2,3). However, these driver mutations mainly cluster in adenocarcinoma. So it is imperative to explore the potential biomarkers for squamous carcinoma.
The phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB/Akt) signaling pathway plays an important role in cell growth and proliferation initiated by activation of receptor tyrosine kinases and in tumor genesis and progression (9,10). The PI3K catalytic alpha (PIK3CA) encodes the p110α catalytic subunit of PI3K, which forms a heterodimer with the p85 regulatory subunit and is sequestered to the cell membrane in response to multiple stimuli. Recently, activating mutations of PIK3CA gene have been identified in a broad spectrum of tumors. A great majority of somatic mutations in PIK3CA are missense mutations clustering in exons 9 and 20 that encode a part of the helical and kinase domains, respectively (11). There is increasing evidence that constitutive activation of the PI3K pathways in lung cancer occurs as a consequence of PIK3CA mutation or amplification. However, in Chinese patients with LSCC, little data exist on PIK3CA mutations.
PIK3CA is a 34 kb gene located on chromosome 3q26.3 that consists of 20 exons coding for 1,068 amino acids yielding a 124 kDa protein (12). PIK3CA gene codes for the catalytic subunit p110α of class IA PI3Ks. The first of these mutational reports was published by Samuels et al. in 2004 (11). Subsequently, PIK3CA mutation is identified in a considerable portion of human primary epithelial cancers, resulting in PIK3CA being one of the most commonly mutated oncogenes in human cancers (13,14). PIK3CA mutations in human cancers confer a gain of function resulting in increased lipid kinase activity and in constitutive activation of the PI3K-AKT pathway in the absence of growth factor (11,15).
The PI3K/Akt pathway lies in downstream of certain receptor tyrosine kinases, including EGFR. However, the reports about occurrence of PIK3CA mutations and its association with EGFR mutation and KRAS mutation in LSCC were limited. The aims of the present study were to investigate PIK3CA mutations in exons 9 and 20 in Chinese patients with LSCC and explore the correlations between PIK3CA mutation and EGFR or KRAS mutations, and the clinicopathologic characteristics.
Materials and methods
Patients and tissue samples
This retrospective study was conducted in a cohort of 123 patients with LSCC who had treated at the Department of Thoracic Oncology of Beijing Cancer Hospital from June 2003 to September 2011. All patients met the following inclusion criteria: (I) having histologically verified LSCC; (II) the formalin-fixed paraffin-embedded (FFPE) tissue having tumor cell no less than 70%; and (III) having basically clinicopathological information. Both adenosquamous lung cancer patients and those with the second primary cancer were excluded. Survival analysis was calculated from date of diagnosis to date of death or last date of contact for those alive at the time of the analysis. This study was approved by the Institutional Ethic Committee of Beijing Cancer Hospital.
FFPE tissues were obtained before therapy. Serial sections (4 μm) containing representative malignant cells were stained with Hematoxylin-Eosin (H&E) and classified based on the World Health Organization criteria. The tumor samples for molecular analysis were obtained from bronchoscopic biopsy or from computed tomography/ultrasound-guided needle biopsy, or from percutaneous aspiration (lymph nodes and skin metastasis), or surgery.
Detection of EGFR and KRAS mutations
The genomic DNA was extracted from FFPE tissues using the EZN AFFPE DNA Kits (OMEGA, USA) according to the manufacturers’ instructions. We performed denaturing high-performance liquid chromatography (DHPLC) using the Transgenomic Wave Nucleic Acid Fragment Analysis System (Transgenomic, Omaha, NE) for EGFR (exon 19 and 21) (16) and KRAS (coden 12 and 13) mutation detection. The temperature for analysis was according to the optimal resolution of heteroduplex and homoduplex determined by analyzing the melting behavior of each fragment. We identified mutation profiles by visual inspection of the chromatograms on the basis of the appearance of earlier-eluting (EGFR 19 product) or blunt peaks (EGFR 21 and KRAS product) and corresponding homozygous profiles showed only one peak.
Detection of PIK3CA mutation by pyrosequencing
PIK3CA mutation testing has been carried out in our laboratory using conventional PCR followed by pyrosequencing. PIK3CA exon 9 is amplified using a forward biotinylated primer, 5'-biotin-CAATGAATTAAGGGAAAATGACAA-3', and a reverse primer, 5'-ACCTGTGACTCCATAGAAAATCTT-3'. PIK3CA exon 20 is amplified using a forward biotinylated primer, 5'-biotin-GCAAGAGGCTTTGGAG TATTTCA-3', and a reverse primer, 5'-GTTCAATGC ATGCTGTTTAATTGT-3'. The PCR master mix contains the forward and reverse primers (each 0.2 μmol/L), 2× mix (Promega, USA) 7.5 μL and 20 ng of sample genomic DNA in a total volume of 15 μL. PCR-cycling conditions for PIK3CA exon 9 consist of initial denaturing at 95 °C for 5 min; 10 cycles of 95 °C for 30 s, 52 °C for 30 s and 72 °C for 50 s; 35 cycles of 95 °C for 30 s, 57 °C for 30 s and 72 °C for 50 s; and final extension at 72 °C for 10 min. PCR-cycling conditions for PIK3CA exon 20 consist of initial denaturing at 95 °C for 5 min; 10 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 50 s; 35 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 50 s; and final extension at 72 °C for 10 min. The reactions were carried out on an ABI 2,720 Thermocycler (Applied Biosystems). The PCR products were electrophoresed in an agarose gel to confirm successful amplifications before pyrosequencing. The PCR products (each 10 μL) are then sequenced by the Pyrosequencing PyroMark Q24 System (Qiagen, Germany), following the manufacturer’s instructions using the pyrosequencing primer for PIK3CA exon 9, 5'-TAGAAAATCTTTCTCCTGC-3', and the pyrosequencing primer for PIK3CA exon 20, 5'-CCATTTTTGTTGTCCAG-3'. Each sample is sequenced with a separate program of nucleotide dispensation orders: 5'-GTCGAGTGCATTCGAGA-3', designed for detecting mutations at exon 9; and 5'-TCGACGTATCGACTGTG-3’ designed for detecting mutations at exon 20. The primers and the dispensation orders enable us to capture all possible mutations of the wild type sequence at exon 9 E542 (GAA), E545 (GAG) and exon 20 H1047 (CAT), G1049 (GGT) of PIK3CA.
We confirmed these cases defined as “mutation positive” by pyrosequencing using Amplification Refractory Mutation System (ARMS) or clone sequencing. PIK3CA mutation was detected using AmoyDxTM PIK3CA Mutation Detection Kit (Amoy Diagnostics Co., LTD, China.) for E542K, E545K, H1047R, H1047L amino acid mutation detection in accordance with the manufacturer’s instructions. For E542Q mutation, clone sequencing was done by selecting 15 sub-clone for each product.
Detection of PIK3CA amplification by fluorescence in situ hybridization
PIK3CA gene copy number of the tumor samples, which had PIK3CA mutations, was also evaluated by fluorescence in situ hybridization (FISH). FFPE tumor materials were cut into 4 μm thick sections and placed onto glass slides. Sections of slides were deparaffinized, pretreated with paraffin pretreatment solution at 95 °C for 30 min and digested with protease solution at 37 °C for 10-20 min. PIK3CA (Texas Red)/CEN3q (FITC) FISH probe (Abnova, Taiwan) was applied to the sections. The sections were denatured at 75 °C for 5 min and then hybridized at 37 °C for 48 h. After hybridization, washing was done. The sections were counterstained with 4’,6-diamidino-2-phenylindole (DAPI). The samples were analyzed under a 100× oil immersion objective with a fluorescence microscope (OLYMPUS, Japan). At least 100 nuclei per case were evaluated. Signal count ratio of PIK3CA to CEP3q more than 2 was defined as amplification.
Statistical analysis
All eligible patients meeting the study inclusion criteria were included in the final statistic analysis. The statistical analyses of categorical variables were done using the Pearson’s χ2 test or the Fisher’s exact test where appropriate. All statistical tests were two sided, and a P value less than 0.05 was considered statistically significant. All statistical procedures were performed with SPSS statistical software, version 16.0 (SPSS Inc., Chicago, IL, USA).
Results
Clinical variables
There were 104 men (84.6%) and 19 women (15.4%), with age at diagnosis ranging from 32 to 83 years (median age, 64.0 years). Patients consisted of 28 non-smokers (22.8%) and 95 smokers (77.2%). Patients had performance status ranging from grade 0 to 3. Performance status was defined according to the Eastern Cooperative Oncology Group. Pathologic staging of lung cancers was determined according to the UICC-AJCC-TNM system (version 7, 2009) (17): 4 (3.3%) had stage I disease, 10 (8.1%) stage II, 19 (15.4%) stage IIIA, 38 (30.9%) stage IIIB, and 52 (42.3%) stage IV. All patients had tissue sample assessable for EGFR mutation, KRAS mutation and PIK3CA mutation detection. The clinicopathologic characteristics of the patients are summarized in Table 1.
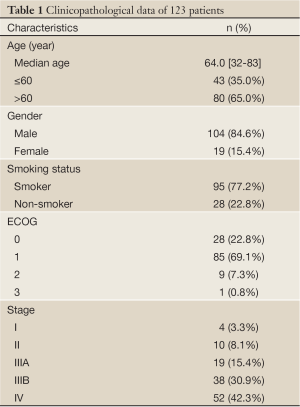
Full Table
PIK3CA mutation in LSCC
Tissues of the 123 patients were available for DNA sequencing analysis. PIK3CA mutations were found in three patients (2.4%), one in the catalytic domain of PIK3CA and two in the helical domain. The mutation included E545K, E452Q, and H1047R. One sample with PIK3CA mutation (E545K) also had EGFR mutation (L858R), the other two coupled with PIK3CA amplification. All the three patients with PIK3CA mutation share the common clinicopathologic characteristics: male, less than 60 years old, had smoke histology, stage III and carried wild-type KRAS (Table 2).

Full Table
EGFR and KRAS mutation in LSCC
The tissues of the 123 patients were available for EGFR mutation analysis. EGFR mutations were found in 19 patients (15.4%), including 9 in exon 19 and 10 in exon 21. The EGFR mutation status was significantly different between smoker and non-smoker groups in our cohort (10.5% vs. 32.1%, P=0.013). The frequency of EGFR mutation was found higher in women than men, but it was not statistically significant (31.6% vs. 12.5%, P=0.077). The incidence of EGFR mutation did not show association with age and stage (Table 3).
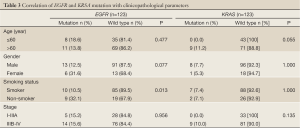
Full Table
The KRAS mutations were identified in 9 (7.3%) of the 123 patients. The KRAS mutations were not correlated with age, gender, smoking status and stage of the LSCC (Table 3).
Discussion
The lung adenocarcinoma genome has been characterized extensively (4-8,18-22). However, less is known about specific genetic alterations, especially “driver mutations”, in LSCC. In present study, we identified 2.4% of Chinese patients with LSCC harboring PIK3CA mutations with the subtype E545K, E452Q and H1047R. The mutational status of PIK3CA is not mutually exclusive to EGFR mutation. All the PIK3CA mutation positive patients shared the same clinicopathologic characteristics, such as male, less than 60 years old, had smoke history, stage III and carried wild-type KRAS.
Although PIK3CA mutations occur frequently in a variety of human cancers, individual types of epithelial cancers show remarkable variability in their mutational rates, with high rates present in colon cancer, breast cancer, gastric cancer, hepatocellular cancer, brain tumors and ovarian cancer (11,14,23-25). Whereas the rates described in NSCLC are relatively uncommon. Samuels et al. reported a low frequency (4.2%) of PIK3CA mutations in lung cancer (11). Kawano and his colleagues then confirmed this low frequency (3.4%) in Japanese lung cancer patients and demonstrated that PIK3CA mutation incidence was more commonly in squamous cell carcinoma (6.5%) than in adenocarcinoma (1.5%) (26). Okudela et al. identified the mutations of PIK3CA in 3.6% of lung cancers (27). Lee et al. detected PIK3CA mutations in 3 of 229 NSCLC (1.3%) (25). Recently, a large samples study was reported. Yamamoto et al. analyzed about 700 lung cancer samples and identified PIK3CA mutations in 1.6% of all major histology types (28). Stratified by squamous cell carcinoma in the above-mentioned studies, the frequency of PIK3CA mutation was 2-7.1%. Our study showed that 2.4% (3/123) of the Chinese LSCC carried mutations in exon 9 and exon 20 of PIK3CA gene, which was in good agreement with previous reports, indicating PIK3CA mutation is also a rare event in Chinese patients with LSCC. Furthermore, the present data confirm that the prevalence of PIK3CA mutations in LSCC doesn’t present ethnic difference, unlike mutations in EGFR (18).
The high number of observed PIK3CA mutations, so-called “hotspot” mutations, map to three sites, E542 and E545 in the helical domain (exon 9) and H1047 in the kinase domain (exon 20). In our study, the three mutated subtypes were observed. In vitro and in vivo oncogenicity of PIK3CA mutants strongly suggests a critical role for these mutated proteins in human malignancies. It was well documented that these mutations cause an increase in PI3K activity and induce oncogenic cellular transformation when expressed in chicken embryo fibroblasts and in mouse NIH3T3 cells (29,30). In the kinase domain, H1047R is the most common mutation that occurs at the end of the activation loop of the p110 α/p85 α complex. Through the use of somatic cell knockouts, Samuels reported that the mutations of PIK3CA result in increased cell signaling, cell growth and invasion. The colorectal cancer cell lines HCT116 containing the hotspot mutation H1047R in exon 20 had a high apoptotic resistance and an increased ability to migrate and metastatize when injected in the tail vein of athymic nude mice (15).
There was one patient with both PIK3CA and EGFR mutation. The mutational status of PIK3CA was not mutually exclusive to EGFR, which is similar to previous reports (26,28,31). It has been reported that PIK3CA mutations were not correlated with gender, smoking status in Japanese lung cancer patients (26). Yamamoto reported that tumor samples with PIK3CA mutation also had KRAS mutations (28). However, in our study all the three patients with PIK3CA mutation share the common clinicopathologic characteristics: male, less than 60 years old, had smoke history, stage III and carried wild-type KRAS. Two possible reasons may result in the discrepancy between our and other studies. Firstly, the discrepancy in tumor histological subtypes exists. Previous studies investigated whole NSCLC, whereas our study focused on analysis LSCC. Secondly, because the occurrence of PIK3CA mutation is a small probabilistic molecular event in LSCC, the positive cases of PIK3CA mutation in both our and other studies were limited, even in the study of Yamamoto et al., there only were 11 patients with PIK3CA mutation, which may contribute to the bias of associations of the PIK3CA mutation with clinicopathologic characteristics and other molecular aberrance.
From the diagnostic and therapeutic standpoint, long-term prospective, blinded randomized trials could be performed to determine if the presence or absence of PIK3CA mutations have any correlation with clinical outcome in LSCC. This would then allow for the clinician to predict with complete certainty whether or not cancers harboring these mutations would be more or less aggressive and could therefore influence decisions for systemic therapies. The development of targeted therapies creates a need to discriminate tumors accurately by their histological and genetic characteristics. Nevertheless we expect that PIK3CA will become a major molecular target for personalized therapy in LSCC.
In summary, PIK3CA mutation occurred in a rare proportion in Chinese patients with primary LSCC, which was similar to Western and Japanese populations. The mutational status of PIK3CA is not mutually exclusive to EGFR mutation. All the patients with PIK3CA mutation shared the same clinicopathologic characteristics: male, less than 60 years old, had smoke history, stage III and carried wild-type KRAS.
Acknowledgements
This work was supported by grants from National Natural Sciences Foundation Distinguished Young Scholars (81025012), National Natural Sciences Foundation General Program (81172235), the Capital Development Foundation (2007-1023), Beijing Health Systems Academic Leader (2011-2-22), and Science and Technology Project of Beijing (Z090507017709015).
Disclosure: The authors declare no conflict of interest.
References
- Youlden DR, Cramb SM, Baade PD. The international epidemiology of lung cancer: geographical distribution and secular trends. J Thorac Oncol 2008;3:819-31. [PubMed]
- Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science 2002;297:63-4. [PubMed]
- Fisher GH, Wellen SL, Klimstra D, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev 2001;15:3249-62. [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [PubMed]
- McLeer-Florin A, Lantuéjoul S. Why technical aspects rather than biology explain cellular heterogeneity in ALK-positive non-small cell lung cancer. J Thorac Dis 2012;4:240-1. [PubMed]
- Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A 2008;105:19893-7. [PubMed]
- Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutation on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer cell 2006;9:485-95. [PubMed]
- Manning BD, Cantley LC. AKT/PKB signalling: navigating downstream. Cell 2007;129:1261-74. [PubMed]
- Fresno Vara JA, Casado E, de Castro J, et al. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 2004;30:193-204. [PubMed]
- Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554. [PubMed]
- Volinia S, Hiles I, Ormondoryd E, et al. Molecular cloning, cDNA sequence, and chromosomal locolization of the human phosphatidylinositol 3-klnase p110 alpha (PlK3CA) gene. Genomics 1994;24:472-7. [PubMed]
- Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer 2006;94:455-9. [PubMed]
- Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol 2006;18:77-82. [PubMed]
- Samuels Y, Diaz LA Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 2005;7:561-73. [PubMed]
- Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009;27:2653-9. [PubMed]
- Edge SB, Byrd DR, Compton CC, et al. eds. AJCC Cancer Staging Manual. 7th ed. New York: Springer, 2010:237-46.
- Hirsch FR, Bunn PA Jr. EGFR testing in lung cancer is ready for prime time. Lancet Oncol 2009;10:432-3. [PubMed]
- Daniels M, Goh F, Wright CM, et al. Whole genome sequencing for lung cancer. J Thorac Dis 2012;4:155-63. [PubMed]
- Eberhard DA, Johnson BE, Amler LC, et al. Mutation in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicator in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Cli Oncol 2005;23:5900-9.
- Miller VA, Riely GJ, Zakowski MF, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol 2008;26:1472-8. [PubMed]
- Tsao MS, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol 2007;25:5240-7. [PubMed]
- Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 2004;3:772-5. [PubMed]
- Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 2004;64:7678-81. [PubMed]
- Lee JW, Soung YH, Kim SY, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 2005;24:1477-80. [PubMed]
- Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer 2006;54:209-15. [PubMed]
- Okudela K, Suzuki M, Kageyama S, et al. PIK3CA mutation and amplification in human lung cancer. Pathol Int 2007;57:664-71. [PubMed]
- Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res 2008;68:6913-21. [PubMed]
- Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA 2005;102:802-7. [PubMed]
- Ikenoue T, Kanai F, Hikiba Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res 2005;65:4562-7. [PubMed]
- Endoh H, Yatabe Y, Kosaka T, et al. PTEN and PIK3CA expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol 2006;1:629-34. [PubMed]


