MiR-182 is up-regulated and targeting Cebpa in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) accounts for over 90% of liver cancers (1), and it has poor prognosis with only about 5% of patients survive more than five years (2). Thus, HCC is one of the most major health problems, with nearly 600,000 deaths each year across the world (3). Therefore, it is worthwhile to investigate the pathogenesis of HCC, so as to develop effective means of prevention and treatment to this severe cancer (4).
MicroRNAs (miRNAs) are small non-coding RNA molecules that can regulate gene expression by specifically recognizing base-pairing sites to their target mRNAs (5). The miRNA system is conserved from worms to mammals (6,7), which indicates its important functions. MiRNAs are involved in many biological processes including cell cycle, differentiation, development, metabolism, and so on (8-13). Recent studies have emphasized the essential roles of miRNAs in diverse diseases (14-16), especially in cancer (17).
Altered miRNA expression is also observed in HCC (18). For instance, miR-21 is dramatically upregulated in HCC tissues and cell lines to increase HCC cell proliferation through targeting PTEN (19) and PDCD4 (20). Another example is that miR-122 is down-regulated in 70% of HCC (21), and an inverse correlation links miR-122 to cyclin G1 in HCC tissues (22).
Due to its essential functions and typical expression pattern, miR-122 has been recognized as the most important HCC biomarker in HCC (23). Previous reports have demonstrated that the expression of miR-122 is positively regulated by CEBPA (24). Then miR-122 represses the translation of IGF1R (insulin-like growth factor 1 receptor) and preserves the activity of GSK3B (glycogen synthase kinase-3 beta). Further GSK3B represses cell proliferation and in feedback promotes the expression of CEBPA. This regulatory circuitry is demonstrated to play important roles in hepatocarcinogenesis. To the best of our knowledge, CEBPA is only reported to be a target of miR-124a in leukemia (25).
In this study, we performed deep sequencing for tumor and adjacent normal tissues of HCC model rat (Morris Hepatoma-3924A). We identified 18 down- and 70 up-regulated miRNAs, with absolute normalized frequency fold change of >2 and at least 1,000 read per ten million transcripts, respectively. In our result, miR-122 is the most severely down-regulated miRNA with log2 fold change smaller than –9, which is consistent with previous reports. We also identified that miR-182 is one of the most dramatically up-regulated miRNAs. Existing studies show that miR-182 plays a role in breast cancer (26) and metastasis of melanoma (27) by targeting FOXO1 (26), FOXO3 and microphthalmia-associated transcription factor (27), respectively. However, the function of miR-182 in HCC is largely unknown until now.
Therefore, we employed four algorithms [Targetscan (28), PITA (29), Hitsensor (30) and Tarbase (31)] to find potential targets of miR-182. We performed GO (Gene Ontology) (32) and PATHWAY enrichment analysis for the predicted targets of miR-182 to investigate the potential function of miR-182 in HCC. These analyses identify that miR-182 is a potential upstream regulatory miRNA of CEBPA. CEBPA (CCAAT enhancer binding protein alpha) is reported to be down-regulated in most cancer diseases, for instance, breast cancer (33) and lung cancer (34).
To verify the upregulation of miR-182 in HCC, we performed quantitative RT-PCR (qRT-PCR) of miR-182, CEBPA and miR-122 in 25 tumor samples and 16 adjacent normal samples of HCC model rat, respectively. The results show significantly increased expression of miR-182, reduced expression of CEBPA, and reduced expression of miR-122 in tumor tissues. Correlation analyses reveal significant negative correlation between the expression of (I) miR-182 and CEBPA; (II) miR-182 and miR-122, and positive correlation between the expression of miR-122 and CEBPA. The luciferase assay experiment validated that Cebpa is a direct target of miR-182 in rat. Because the miR-182 complementary site in the 3' untranslated region (UTR) of CEBPA is highly conserved in vertebrates, we further verified that miR-182 also directly targets CEBPA in human. These results verify that miR-182 is an upstream regulator of the established CEPBA/miR-122/GSK3B pathway that regulates the proliferation of tumor cells. These findings suggest that miR-182 could play a role of oncogene in HCC and potentially could be used as a biomarker for the diagnosis and treatment of HCC.
Material and methods
Ethics statement
The housing facilities of animal models is a barrier housing facilities, and it has in keeping with national standard “Laboratory Animal-Requirements of Environment and Housing Facilities” (GB14925-2001). The care of laboratory animal and the animal operation conform to national and Shanghai municipality’s regulations for the administration of affairs concerning experimental animals. The protocol was approved by Institutional Animal Care and Use Committee of Zhongshan Hospital of Fudan University (Permit Number: SYXK 2008-0039).
All surgery was performed under ketamine anesthesia, and all efforts were made to minimize suffering.
Collection of experimental samples
A total of 30 male ACI rats with weight 250-300 g (average of 280 g) from the Liver Cancer Institute of Fudan University were performed with liver cancer transplantation (tumor strain: Morris Hepatoma-3924A) successfully. The HCC samples and the adjacent normal (NT) samples were collected two weeks after tumor transplantation. The main steps included: (I) the rats were anesthetized by intraperitoneal injection of ketamine (100 mg/kg); (II) the liver was exposed and the HCC tissues and NT tissues, about 0.5 cm3, from the liver were taken; (III) all the samples were put into the liquid nitrogen tank immediately; and (IV) the animals were sacrificed by cervical dislocation.
RNA extraction
Tissues were then cut into cubes of about 0.5 cm3 and kept in liquid nitrogen. RNA was extracted from the frozen tissues using Trizol reagent (Invitrogen) according to the manufacturer’s instructions, and qualified with Agarose gel electrophoresis. Unqualified RNA samples were excluded in subsequent experiments. We finally obtained 24 tumor and 16 adjacent normal tissue RNA for further use, of which 14 tumor and 14 adjacent normal tissues were paired.
Deep sequencing
One pair of tumor and adjacent tissues were delivered for deep sequencing using Illumina HiSeq2000 Sequencer. The quality of two RNA samples was further verified using ultraviolet spectrophotometry and 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). Deep sequencing was performed by following standard protocols.
Analysis of small RNA sequencing libraries
We analyzed the small RNA libraries as previously reported (35,36). After removing the 3' adapters of reads and discarding reads with low qualities, we obtained about 20.8 million reads with 18 to 45 nucleotides, representing around 983,632 million unique sequences. All the sequences were aligned with mature miRNAs, pre-miRNAs, mRNAs, ncRNA, repeat elements and genome with SOAP2 (37), allowing no mismatches. Mature miRNAs and pre-miRNAs sequences were downloaded from the miRBase (r19, http://mirbase.org). The whole genome and mRNA sequences were downloaded from UCSC Genome Browser (http://genome.ucsc.edu/). The ncRNA sequences were downloaded from the Rfam (http://rfam.sanger.ac.uk/), NONCODE (http://noncode.org), Genomic-tRNA-db (http://gtrnadb.ucsc.edu), and Silva (http://www.arb-silva.de) databases. The repeat element sequences were downloaded from Repbase (http://www.girinst.org/repbase/).
The expression profiles of miRNAs were obtained with reads that had no mismatches to mature rat miRNAs and then were normalized to reads per ten million transcripts (RPTM).
miRNA target prediction
Mature sequences of miRNAs were downloaded from miRBase and the 3' UTR sequence of rat Cebpa and human CEBPA were downloaded from UCSC Genome Browser. The hsa-miR-182 targets predicted by at least two of the four algorithms, Targetscan (28), PITA (29), Hitsensor (30) and Tarbase (31), or experimentally verified from miR2Disease (38), miRecords (39) and TarBASE (31) were combined and used to perform GO enrichment analysis.
GO enrichment analysis of the targets of hsa-miR-182
We conducted GO enrichment analysis for three categories of GO: biological process, cellular component and molecular function separately using the same method. The P-value used for GO enrichment analysis was defined as Eq.[1]:

where m was the number of genes in the considering GO term, n was the total number of genes in all the GO terms, k was the number of target genes of miR-182, and c was the number of target genes of miR-182 which belonged to the considering GO term.
Pathway enrichment analysis of the targets of hsa-miR-182
Similarly the data downloaded from PathwayAPI database (40) was utilized to evaluate the pathway enrichment for target genes of miR-182. Three most widely used pathway databases KEGG (http://www.genome.ad.jp/kegg), Ingenuity (http://www.ingenuity.com/), and Wikipath-ways (http://www.wikipathways.org/), were combined in the PathwayAPI database. We evaluated the pathway enrichment for target genes of miR-182 based on the P-value of the hypergeometric test in Eq. [1], where m denoted the number of genes in the considering pathway, n was the total number of genes in all the pathways, k referred to the number of target genes of miR-182, and c was the number of target genes of miR-182 which belonged to the considering pathway.
miRNA real-time PCR analysis
For candidate miRNAs, the reverse transcriptions were carried out using High Capacity RNA-to-cDNA kit (Takara) with no more than 500 ng RNA per 10 µL PCR system as template. qRT-PCR was then taken using TaqMan probe (Roche). Amplification of miRNAs was carried out using the TaqMan probe and Mater Mix (HaoQin) in Roche Light Cycler 480 Real-time PCR system. Gene expression was presented relative to RNU6B gene.
Real-time PCR for mRNA expression
Real-time PCR to measure the mRNA expression of CEBPA was carried out using SYBR green detection and standard techniques. cDNA was transcribed from total RNA using random 9 primers. PCR primers were designed by MacVector according to the gene sequence, and had been verified. Gene expression was presented relative to β-actin.
Plasmid construction
To generate the specific miRNA expression vector (pSuper-hsa-miR-182 and pSuper-rno-miR-182), we amplified about 400 base-pair genomic regions containing the miRNA precursor sequences, using primers as listed in Supplementary Figure S1. The amplified regions were then purified and introduced into pSuper vector by restriction sites BglII and HindIII, and were verified by DNA sequencing. The putative miR-182 binding site in the 3' UTR of target gene (human CEBPA and rat Cebpa, respectively) were cloned into pGLO (E1330, Promega, Madison, WI, USA) vector downstream of firefly luciferase 3' UTR as a primary luciferase signal with rellina luciferase as the normalization signal. The pGLO vector itself provided renilla luciferase signal as normalization to compensate the differences between transfection and harvested efficiencies.
Luciferase reporter assay
293T cells were grown in DMEM containing 10% FBS, 5% glutamine and 100 µg/mL penicillin streptomycin. For luciferase reporter assay, co-transfection was performed with 5 µg of either pSuper-rno-miR-182 (pSuper-has-miR-182) or empty pSuper vector, 300 ng of either pGLO-rno-Cebpa (pGLO-hsa-CEBPA) or empty pGLO vector, using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instruction. Total RNA and protein were collected 48 h after transfection and analyzed using the Dual-Luciferase reporter assay System (Promega, Madison, WI, USA). Each transfection had three replicates and was repeated in three independent experiments.
Results
Abundance of miRNAs obtained from hight-throughput sequencing
To obtain miRNAs that are differentially expressed between tumor and adjacent normal tissues in HCC, we perform high throughput sequencing for one pair of tumor and adjacent normal tissue, respectively, using Illumina HiSeq 2000 sequencer. We obtain 20.8 million reads within 18 to 45 nucleotides, representing 983,632 unique sequences. These reads are aligned to different categories allowing no mismatches, including pre-miRNAs, non-coding RNAs (tRNA, rRNA, snRNA, snoRNA, piwiRNA, etc.), mRNAs, repeat elements, and genome (see Materials and Methods). As shown in Figure 1A, miRNAs account for about 40% of the obtained reads in both libraries. However, the percentage of unique sequences that are matched to miRNAs only contributes to about 2% of total unique sequences, suggesting miRNAs are highly expressed in all small RNAs (Figure 1B).
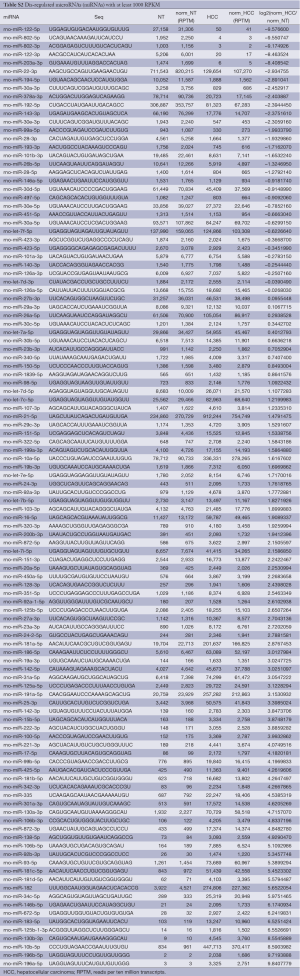
The deep sequencing reads are mapped to 723 rat miRNAs from miRBase (41) allowing no mismatches and then normalized to RPTM to estimate the expression levels of miRNAs. A total of 452 miRNAs have at least one read in our sequencing profiles (Table S1). The most abundant miRNAs include miR-21-5p, miR-22-3p, miR-192-5p, miR-10b-5p, miR-10a-5p, and let-7f-5p as shown in Figure 1C.

Full table
To identify dis-regulated miRNAs between HCC and normal tissues, the log2 ratios of miRNAs with at least 1,000 RPTM in at least one tissue are calculated for 112 miRNAs. Then we find 18 and 70 miRNAs that show reduced and increased expression levels (with log2 ratios 1), respectively, in HCC tissues (Table S2). From Figure 1D and Table 1 (with references in Table S3, 12 miRNAs are severely down-regulated in HCC tissues with more than four fold changes. miR-122 is the most severely down-regulated miRNA, which is consistent with most previous studies (21,24). A total of 51 miRNAs are upregulated by at least four fold (Table S2. Among these 51 upregulated miRNAs, miR-182 is reported to promote breast cancer genesis by targeting FOXO1 (26,42), and metastasis of melanoma by targeting FOXO3 and microphthalmia-associated transcription factor (27).
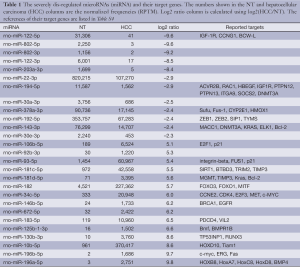
Full table
Full table
Full table
GO and pathway analysis for the targets of miR-182
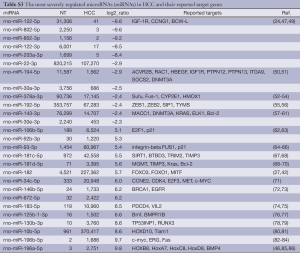
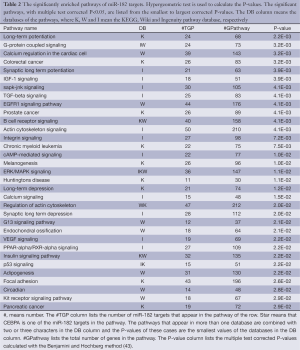
Among the top 20 most severely up-regulated miRNAs, we focus our attention on miR-182, because it is previously reported to be responsible for tumor genesis in other cancers with increased expression levels (26,27,42). However, the mechanism how miR-182 is involved in HCC is not clear. As miRNAs function through its target genes, we employed four miRNA target prediction algorithms including Targetscan (28), PITA (29), Hitsensor (30) and Tarbase (31) to predict hsa-miR-182 target genes. Only targets predicted by at least two of these four algorithms are kept as reliable candidates. We also collect experimentally verified targets of hsa-miR-182 from miR2Disease (38), miRecords (39) and TarBASE (31). The predicted and experimentally verified targets of hsa-miR-182 are combined and used to perform GO and Pathway enrichment analysis. As shown in Table 2, hsa-miR-182 is involved in G-protein-couples signaling, IGF-1 signaling and EGFR1 signaling pathway. Besides, Table 2 show that hsa-miR-182 is also involved in four cancers including colorectal cancer (P=3.2×10–3, hypogeometric test), prostate cancer (P=4.1×10–3, hypogeometric test), chronic myeloid leukemia (P=7.5×10–3, hypogeometric test) and pancreatic cancer (P=2.9×10–2, hypogeometric test). The enriched GO terms of miR-182 targets include nerve growth factor receptor signaling pathway (P=1.6×10–2, hypogeometric test) and protein binding (P=6.9×10–16, hypogeometric test) (Table S4).

Full table
In our prediction, CEBPA is one of targets involved in EGFR signaling pathway and Adipogenesis (Table 2). Besides, previous study reported a regulatory pathway in HCC with CEBPA as upstream regulator targeting hsa-miR-122 (24). Therefore we next investigate whether hsa-miR-182 influences this pathway by targeting CEBPA.
Full table
Validation of miR-182, Cebpa, miR-122 expression in rat samples by qRT-PCR
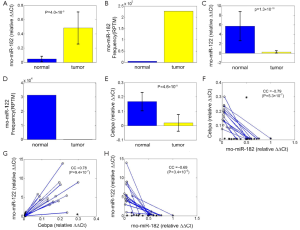
Expression levels of miR-182 and Cebpa are both estimated in 24 adjacent tumor tissues and 16 HCC tissues respectively. From Figure 2A, miR-182 is significantly up-regulated in HCC tissues (P=4.0×10–9, t-test), which is consistent with its sequencing result (Figure 2B). And miR-122 is down-regulated in HCC tissues (P=1.3×10–10, t-test, Figure 2C), consistent with sequencing result in Figure 2D. Cebpa is significantly down-regulated in HCC tissues (P=4.6×10–9, t-test, Figure 2E).
Moreover, we plot the relative expression level of miR-182 and Cebpa of 24 adjacent tumor tissues and 16 HCC tissues (of which 14 are paired HCC and NT tissue samples) in Figure 2F. Their expression levels show significant negative correlation (CC =–0.79, P=5.3×10–7, t-test), which is also consistent with the assumption that Cebpa is a potential target of miR-182. The expression levels of Cebpa:miR-122 pair and miR-182:miR-122 pair show significant positive (CC =0.78, P=9.4×10–7, t-test, Figure 2G) and negative correlation (CC =–0.69, P=3.4×10–5, t-test, Figure 2H), respectively. The correlation analysis reveals that miR-182 should be a potential upstream regulator of CEBPA/miR-122/GSK3B regulatory circuitry. Next we use luciferase assay to verify direct repression of Cebpa by miR-182.
Cebpa is one direct target of miR-182 in rat
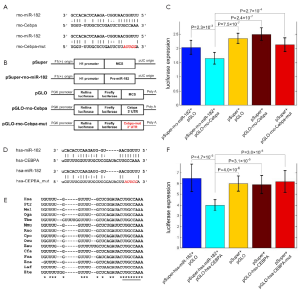
The prediction results reveal that Cebpa is a potential target of miR-182 with an 8 mer seed complementary site (Figure 3A). To investigate the direct relationship between miR-182 and Cebpa, we employ the dual-luciferase reporter system. The 3' UTR region of Cebpa containing the wild-type and mutated miR-182 complementary site are conducted into luciferase reporter plasmid (pGLO), respectively. HEK 293T cells co-transfected with pSuper-rno-miR-182 plasmid and pGLO-rno-Cebpa plasmid show a significantly decreased luciferase expression level compared to control groups (P=2.3×10–3, 7.5×10–7, and 2.4×10–7, t-test, for pSuper-rno-miR-182 + pGLO, pSuper + pGLO, pSuper + pGLO-rno-Cebpa, respectively, Figure 3B). And the luciferse expression levels of cells transfected with pGLO-rno-Cebpa-mut are significantly higher than the experiment group (P=2.7×10–4, t-test, Figure 3C), indicating that miR-182 can directly repress Cebpa by targeting the complementary site in Figure 3A.
CEBPA is also a direct target of miR-182 in human
Most rat miRNAs are highly conserved between rat and human (44), implying their essential functions. The miR-182 complementary site in the 3' UTR of human CEBPA (in Figure 3D) is highly conserved in vertebrates, including rat and human, as shown in Figure 3E. Similarly, we use dual-luciferase reporter assay to verify whether human miR-182 also directly represses the expression of CEBPA. HEK 293T cells co-transfected with pSuper-hsa-miR-182 plasmid and pGLO-hsa-CEBPA show a dramatically reduced luciferase expression level compared to other control groups (P=4.7×10–5, 4.0×10–6, and 3.1×10–5, t-test, for pSuper-hsa-miR-182 + pGLO, pSuper + pGLO, pSuper + pGLO-hsa-CEBPA, respectively, Figure 3F). Similarly, the luciferase expression levels of cells transfected with pGLO-hsa-CEBPA-mut are also not affected by miR-182 expression (P=3.0×10–5, t-test, Figure 3F), similar to the results of rno-miR-182 and Cebpa. In summary, these results demonstrate that miR-182’s direct repression on CEBPA is conserved in human.
Discussion
In this study we use deep sequencing method to obtain the miRNA expression profiles of HCC and adjacent normal tissues. Our results identify many dis-regulated miRNAs, including many reported HCC-related miRNAs, such as miR-21 and miR-122. For example, in our sequencing data miR-196a-5p and miR-196b-5p are the most up-regulated miRNAs. miR-196 is reported to be involve in some human cancers with higher expression levels (45,46). The well investigated HCC biomarker miR-122, which is down-regulated in more than 70% HCC patients (47), also shows a dramatic reduced expression level in our rat HCC tissues.
To predict the potential functions of miR-182, we perform Gene Ontology and Pathway enrichment analysis for its targets. The results show that miR-182 is involved in multiple cancer pathways (Table 2). Previous report has demonstrated that miR-182 can control the expression of FOXO1 (26) in breast cancer to influence tumorigenesis and FOXO3 (27) to promote metastasis of melanoma. However, the importance of miR-182 has not been noticed in HCC.
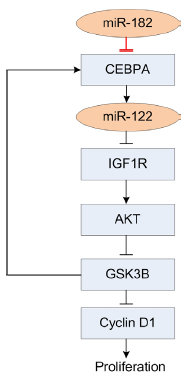
In this study, we verify that miR-182 directly targets Cebpa through a conserved complementary site in both rat and human. A previous report finds that the CEBPA/miR-122/IGF1R pathway is related with proliferation of HCC cells (24). In this pathway, miR-122 is positively regulated by CEBPA. Then miR-122 downregulates the translation of IGF1R and preserves the activity of GSK3B. Further GSK3B represses cell proliferation and in feedback promotes the expression of CEBPA. This regulatory pathway is demonstrated to play important roles in hepatocarcinogenesis (24). In this study, we find that miR-182 is also an important player of this pathway by directly repressing CEBPA (Figure 4). Our results show that the expression levels of miR-182 and Cebpa, miR-182 and miR-122, Cepba and miR-122 are significantly correlated (CC =–0.79, –0.69, and 0.78, P=5.3×10–7, 3.4×10–5, and 9.4×10–7, respectively, t-test, Figure 2F,H,G respectively).
In addition, CEBPA, HNF1A (hepatocyte nuclear factor 1-alpha), HNF3B, HNF4A also activates miR-122 during liver development (48). miR-122 subsequently represses CUTL1 (cut-like homeobox 1) (48). These four transcription factors and miR-122 thus cooperate to maintain the balance of differentiation and proliferation in hepatocytes (48). Our results suggest that miR-182 is potentially involved in liver development as well.
In summary, we find that miR-182 is significantly up-regulated in rat HCC tissues. Our results also verify that miR-182 regulates the established CEBPA/miR-122/IGF1R pathway in our rat HCC models by directly repressing Cebpa. We also verify that miR-182 represses CEBPA in human through a conserved complementary site. These indicate that miR-182 is an upstream regulator of Cebpa in both rat and human. Because miR-122 has been recognized as a key regulator in HCC, our results suggest that miR-182 is a potential biomarker for diagnosis and therapeutic target of HCC. In addition, our results suggest that miR-182 might be involved in several other cancers (see Table 2).
Acknowledgements
The research was supported in part by a start-up grant of Kunming University of Science and Technology given to Yun Zheng, a major projects of Shanghai Municipal Health Bureau (No. 20100222) given to Xiaolin Wang, and a Youth Fund of Zhongshan Hospital, Fudan University (No. 201102) given to Chenggang Wang.
Disclosure: The authors declare no conflict of interest.
References
- Yang ZF, Ho DW, Ng MN, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008;13:153-66. [PubMed]
- Hao K, Luk JM, Lee NP, et al. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer 2009;9:389. [PubMed]
- Liu Y, Wu F. Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ Health Perspect 2010;118:818-24. [PubMed]
- Wang J, Xiong X. Current situation and perspectives of clinical study in integrative medicine in china. Evid Based Complement Alternat Med 2012;2012:268542.
- Ambros V. microRNAs: tiny regulators with great potential. Cell 2001;107:823-6. [PubMed]
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [PubMed]
- Comai L, Zhang B. MicroRNAs: key gene regulators with versatile functions. Plant Mol Biol 2012;80:1. [PubMed]
- Lai VK, Ashraf M, Jiang S, et al. MicroRNA-143 is a critical regulator of cell cycle activity in stem cells with co-overexpression of Akt and angiopoietin-1 via transcriptional regulation of Erk5/cyclin D1 signaling. Cell Cycle 2012;11:767-77. [PubMed]
- Ng R, Song G, Roll GR, et al. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest 2012;122:1097-108. [PubMed]
- Trompeter HI, Dreesen J, Hermann E, et al. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics 2013;14:111. [PubMed]
- Tong MH, Mitchell DA, McGowan SD, et al. Two miRNA clusters, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol Reprod 2012;86:72. [PubMed]
- Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 2012;13:239-50. [PubMed]
- Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011;469:336-42. [PubMed]
- Lu M, Zhang Q, Deng M, et al. An analysis of human microRNA and disease associations. PLoS One 2008;3:e3420. [PubMed]
- Chen X, Liu MX, Yan GY. RWRMDA: predicting novel human microRNA-disease associations. Mol Biosyst 2012;8:2792-8. [PubMed]
- van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov 2012;11:860-72. [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [PubMed]
- Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006;25:2537-45. [PubMed]
- Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647-58. [PubMed]
- Yao Q, Xu H, Zhang QQ, et al. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun 2009;388:539-42. [PubMed]
- Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem 2006;99:671-8. [PubMed]
- Fornari F, Gramantieri L, Giovannini C, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res 2009;69:5761-7. [PubMed]
- Bihrer V, Friedrich-Rust M, Kronenberger B, et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol 2011;106:1663-9. [PubMed]
- Zeng C, Wang R, Li D, et al. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology 2010;52:1702-12. [PubMed]
- Hackanson B, Bennett KL, Brena RM, et al. Epigenetic modification of CCAAT/enhancer binding protein alpha expression in acute myeloid leukemia. Cancer Res 2008;68:3142-51. [PubMed]
- Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 2009;284:23204-16. [PubMed]
- Segura MF, Hanniford D, Menendez S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A 2009;106:1814-9. [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [PubMed]
- Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet 2007;39:1278-84. [PubMed]
- Zheng Y, Zhang W. Animal microRNA target prediction using diverse sequence- specific determinants. J Bioinform Comput Biol 2010;8:763-88.
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA 2006;12:192-7. [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [PubMed]
- Gery S, Tanosaki S, Bose S, et al. Down-regulation and growth inhibitory role of C/EBPalpha in breast cancer. Clin Cancer Res 2005;11:3184-90. [PubMed]
- Halmos B, Huettner CS, Kocher O, et al. Down-regulation and antiproliferative role of C/EBPalpha in lung cancer. Cancer Res 2002;62:528-34. [PubMed]
- Zhang X, Zheng Y, Jagadeeswaran G, et al. Identification and developmental profiling of conserved and novel microRNAs in Manduca sexta. Insect Biochem Mol Biol 2012;42:381-95. [PubMed]
- Jagadeeswaran G, Nimmakayala P, Zheng Y, et al. Characterization of the small RNA component of leaves and fruits from four different cucurbit species. BMC Genomics 2012;13:329. [PubMed]
- Li R, Yu C, Li Y, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 2009;25:1966-7. [PubMed]
- Jiang Q, Wang Y, Hao Y, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 2009;37:D98-104. [PubMed]
- Xiao F, Zuo Z, Cai G, et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 2009;37:D105-10. [PubMed]
- Soh D, Dong D, Guo Y, et al. Consistency, comprehensiveness, and compatibility of pathway databases. BMC Bioinformatics 2010;11:449. [PubMed]
- Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140-4. [PubMed]
- Kim KM, Park SJ, Jung SH, et al. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 2012;27:1669-79. [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289-300.
- Lee CT, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol 2007;26:209-18. [PubMed]
- Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 2007;26:4442-52. [PubMed]
- Schimanski CC, Frerichs K, Rahman F, et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol 2009;15:2089-96. [PubMed]
- Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007;67:6092-9. [PubMed]
- Xu H, He JH, Xiao ZD, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 2010;52:1431-42. [PubMed]
- Lin CJ, Gong HY, Tseng HC, et al. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun 2008;375:315-20. [PubMed]
- Senanayake U, Das S, Vesely P, et al. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis 2012;33:1014-21. [PubMed]
- Meng Z, Fu X, Chen X, et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology 2010;52:2148-57. [PubMed]
- Lee DY, Deng Z, Wang CH, et al. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A 2007;104:20350-5. [PubMed]
- Mohri T, Nakajima M, Fukami T, et al. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol 2010;79:1045-52. [PubMed]
- Skrzypek K, Tertil M, Golda S, et al. Interplay between heme oxygenase-1 and miR-378 affects non-small cell lung carcinoma growth, vascularization, and metastasis. Antioxid Redox Signal 2013;19:644-60. [PubMed]
- Krupa A, Jenkins R, Luo DD, et al. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 2010;21:438-47. [PubMed]
- Boni V, Bitarte N, Cristobal I, et al. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther 2010;9:2265-75. [PubMed]
- Zhang Y, Wang Z, Chen M, et al. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol Cancer 2012;11:23. [PubMed]
- Ng EK, Tsang WP, Ng SS, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer 2009;101:699-706. [PubMed]
- Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009;28:1385-92. [PubMed]
- Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009;460:705-10. [PubMed]
- Zhang H, Cai X, Wang Y, et al. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep 2010;24:1363-9. [PubMed]
- Li Y, Tan W, Neo TW, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci 2009;100:1234-42. [PubMed]
- Ivanovska I, Ball AS, Diaz RL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 2008;28:2167-74. [PubMed]
- Fang L, Deng Z, Shatseva T, et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene 2011;30:806-21. [PubMed]
- Du L, Schageman JJ, Subauste MC, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res 2009;7:1234-43. [PubMed]
- Kan T, Sato F, Ito T, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology 2009;136:1689-700. [PubMed]
- Schonrock N, Humphreys DT, Preiss T, et al. Target gene repression mediated by miRNAs miR-181c and miR-9 both of which are down-regulated by amyloid-β. J Mol Neurosci 2012;46:324-35. [PubMed]
- Wang B, Hsu SH, Majumder S, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 2010;29:1787-97. [PubMed]
- Zhang W, Zhang J, Hoadley K, et al. miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol 2012;14:712-9. [PubMed]
- Wang XF, Shi ZM, Wang XR, et al. MiR-181d acts as a tumor suppressor in glioma by targeting K-ras and Bcl-2. J Cancer Res Clin Oncol 2012;138:573-84. [PubMed]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ 2010;17:193-9. [PubMed]
- Garcia AI, Buisson M, Bertrand P, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med 2011;3:279-90. [PubMed]
- Katakowski M, Zheng X, Jiang F, et al. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest 2010;28:1024-30. [PubMed]
- Li J, Fu H, Xu C, et al. miR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer 2010;10:354. [PubMed]
- Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett 2008;582:3663-8. [PubMed]
- Xia HF, He TZ, Liu CM, et al. MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem 2009;23:347-58. [PubMed]
- Saetrom P, Biesinger J, Li SM, et al. A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res 2009;69:7459-65. [PubMed]
- Ma S, Tang KH, Chan YP, et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 2010;7:694-707. [PubMed]
- Lai KW, Koh KX, Loh M, et al. MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer 2010;46:1456-63. [PubMed]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007;449:682-8. [PubMed]
- Moriarty CH, Pursell B, Mercurio AM. miR-10b targets Tiam1: implications for Rac activation and carcinoma migration. J Biol Chem 2010;285:20541-6. [PubMed]
- Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem 2010;340:97-106. [PubMed]
- Coskun E, von der Heide EK, Schlee C, et al. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res 2011;35:208-13. [PubMed]
- Li Z, Huang H, Chen P, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun 2012;3:688. [PubMed]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004;304:594-6. [PubMed]
- Braig S, Mueller DW, Rothhammer T, et al. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci 2010;67:3535-48. [PubMed]



