Kaiso mainly locates in the nucleus in vivo and binds to methylated, but not hydroxymethylated DNA
Introduction
Kaiso (ZBTB33) is a ubiquitously expressed BTB/POZ transcriptional factor that has firstly been identified as an interaction partner of p120-catenin in the cytoplasm (1). Kaiso is also found to specifically bind with clusters of methylated-CGCG sequences and unmethylated Kaiso binding site TCCTGCNA (KBS) in vitro (2,3). Both three zinc fingers and their flanking domains in Kaiso might contribute to the CGCG binding (4). Although Blattler et al. have reported that Kaiso may bind with transcriptional active genes in the genome of cell lines (5), it is still believed that Kaiso represses its target genes through binding to the CGCG-containing and KBS sequences (6-8). Transcriptional repression by Kaiso may play roles in the development and carcinogenesis (9,10).
As an intermediate contributing factor to active DNA demethylation, 5-hydroxymethyl-cytosine (hmC) plays a vital role in stem cell biology and lineage-specific differentiation (11-14). Interaction protein of hmC that can mediate its transcriptional regulation has been well studied (15-18). These proteins include methyl-CpG-binding domain (MBD) proteins MBD1, MBD2, MBD3, MBD4, and MeCP2. Only MBD3 and MeCP2 showed binding affinity to hmC (19). Although there is a report indicating that Kaiso cannot bind to KBS site containing hydroxymethylation (20), Kaiso’s affinity to hydroxymethylated-CGCG sequence has not been examined.
In addition, reports on the subcellular localization of Kaiso are conflicting. Different subcellular localizations of Kaiso were observed between cultured cell lines and tissues (21). Two recent studies revealed a new target for Kaiso at the centrosomes and spindle microtubules during mitosis, providing a new interpretation of the presence of cytoplasmic Kaiso (22,23). Kaiso contains one nuclear localization signal (NLS) (24). It was suggested that p120-catenin might play a role in Kaiso’s translocation within cells (25,26). That only the nucleic Kaiso correlates with invasion or prognosis of cancers indicates the importance of its intracellular translocation in carcinogenesis (27).
In most of studies, Kaiso was detected by means of indirect immunochemical staining, thus non-specific staining could not completely be excluded. Therefore, we initially compared the subcellular location states of Kaiso in cancer cells (in vitro and in vivo) and found that Kaiso was mainly located in the nucleus in vivo. Most importantly, we found that nuclear Kaiso could not bind with the hydroxymethylated-CGCG sequence in the present study.
Materials and methods
Plasmid constructions
The full-length Kaiso coding sequence (Gene Symbol: ENSG00000177485) was amplified from human cDNA with primers (sense 5’-attaaactcg aggcatggag agtagaaaac tga-3’ and antisense 5’-cgcttcgaat tcgtttagta agactctggt attat-3’), then inserted between Xho I and EcoR I sites in pEGFP-C1 vector to generate pEGFP-C1-Kaiso. To construct pGEX-4T-1-Kaiso, POZ domain deletion Kaiso was amplified with primers sense 5’-tatcggaatt ccctgactcg gccgtcagt-3’ and antisense 5’-attgcctcga gcattggctt gttctgagt-3’, then inserted into EcoR I and Xho I sites in vector pGEX-4T-1 as previously described (4).
Cell culture, transfection, and stable cell line generation
Gastric cancer cell line MGC803 (28), kindly provided by Dr. Yang Ke at Peking University Cancer Hospital & Institute in 2004, was grown in RPMI-1640 medium (Gibco). Human colon cancer cell line HCT116, kindly provided by Dr. Yuanjia Chen, Peking Union Hospital (ATCC CCL-247), was cultured in DMEM (Gibco). The media were supplemented with 10% heat-inactivated fetal bovine serum (Gibco) plus antibiotics. Both cell lines were tested and authenticated by Beijing JianLian Gene Technology Co., Ltd. before they were used in this study. Short tandem repeat (STR) patterns were analyzed using GoldeneyeTM20A STR Identifiler PCR Amplification Kit.
MGC803 cells were transfected with pEGFP-C1 and pEGFP-C1-Kaiso respectively using Lipofectamine 2000 reagent (Invitrogen) following provider’s suggestion. Cells transfected with pEGFP-C1 or pEGFP-C1-Kaiso were selected with 750 µg/mL of G418 (Sigma) for 2 weeks, and sorted by flow cytometry (BD FACS Aria) for subsequent studies.
In vivo cell suspension generation, ex vivo culture, confocal microscopy examination
MGC803 cells (2×106) stably expressing enhanced green fluorescent protein (EGFP) or EGFP-Kaiso in 0.2 mL phosphate buffered saline (PBS) were inoculated intraperitoneally (i.p.) into the peritoneum of 6 week-old male BALB/c nude mice, respectively. These mice were sacrificed 21 d after inoculation. The cell suspension in the peritoneum was collected using PBS washes. The in vitro cultured cells and cells in peritoneum suspension were rinsed three times with PBS, fixed with 4% paraformaldehyde in PBS for 10 min at 37 °C, washed three times in PBS, counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (1 µg/mL) for 3 min, and then examined with Leica SP5 Laser Scanning Confocal Microscopy.
Immunohistochemistry staining (IHC)
Mouse anti-Kaiso monoclonal antibody (1:100; Abcam, ab12723) was used in IHC analysis to detect expression status of Kaiso protein in gastric carcinoma tissues according to the instruction. Regular mouse IgG was used as negative control. The representative gastric carcinoma tissue samples were from inpatients with surgical treatment at Peking University Cancer Hospital under the approved institutional guidelines. All patients gave a written informed consent.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot assays
RIPA buffer lysates of MGC803 cells were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. The membrane was then blocked with 5% skim milk in PBST and probed with indicated antibodies according to the manual instruction. For Western blot detection, we utilized rabbit anti-EGFP and mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies (Proteintech Group), horseradish-peroxidase (HRP)-conjugated goat anti-rabbit secondary or anti-mouse secondary antibodies (Zhongshan Goldenbridge). Protein bands were further detected with HRP Substrate Luminol Reagent (Millipore).
Electrophoretic mobility shift assay (EMSA)
A CDKN2A (p16ink4a) promoter-specific PCR primer set (sense 5’-gaacgcactc aaacacgcc-3’ and antisense 5’-gttggcaagg aaggaggact-3’) was used to prepare the CGCG-containing probe (115 bp) through PCR. 5-Methyl (or 5-hydroxymethyl) dCTP (Bioline) was utilized to synthesize the methylated (mC) [or hydroxymethylated (hmC)] probe. All the probes were biotin-labeled according to the instruction of Biotin 3’ End DNA Labeling Kit (Thermo). The unlabeled probes were used as competitive probes in the competitive EMSA assay.
For purification of GST-Kaiso and GST, sonicated lysates of isopropyl β-D-1-thiogalactopyranoside (IPTG)-induced GST-Kaiso or GST from Escherichia coli (E. coli) were incubated with glutathione-sepharose beads (GE healthcare). After washing with PBS, the beads were incubated with elution buffer (50 mmol/L Tris-HCl pH 8.0, 10 mmol/L GSH-R) for 1 h at 4 °C. The purities were tested both by Coomassie Brilliant blue staining and Western blot probed with anti-GST antibodies (Proteintech Group).
Biotin-labeled probes were respectively incubated with the purified GST-Kaiso and GST proteins in the binding buffer [25 mmol/L HEPES pH7.5, 100 mmol/L KCl, 1 mmol/L ethylene diamine tetraacetic acid (EDTA), 10 mmol/L MgCl2, 0.1% NP40, 5% glycerol and 1 mmol/L dithiothreitol (DTT)] on ice for 1 h. For competitive or antibody reaction, 10 times of unlabeled probe or anti-Kaiso antibody (Abcam) was pre-incubated with the proteins for 30 min, then biotin-labeled probe was added into the reaction for 30 min. Each reaction was loaded onto a 4% polyacrylamide gel in 0.5× TBE (45 mmol/L Tris borate, 1 mmol/L EDTA) and electrophoresed at 100 V for 1.5 h, then transferred onto nylon membrane (Amersham Biosciences) at 80 V for 1.5 h. The binding of the biotin-labeled probes was detected according to Chemiluminescent Nuclear Acid Detection Module (Thermo).
Chromatin immunoprecipitation assay (ChIP)
The ChIP assay was performed as previously reported with slight modifications (29). DNA samples were purified using DNA purification and concentrator kit (Zymo research) using 30 µL for the final elution from the column. About 500 ng of input or Kaiso precipitated DNA was divided into two aliquots. One was used for glucosylation treatment with 4 units of T4 β-glucosyltransferase (T4-β-GT, Zymo Research), the other was used for glucosylation negative control without addition of T4-β-GT.
Both Msp I and Hpa II digest the unmethylated CCGG sites. Msp I also digests methylated- and hydroxymethylated-CCGGs (CmCGG and ChmCGG), but cannot digest glucosylated-ChmCGG (CghmCGG); while Hpa II cannot digest any modified CCGG such as CmCGG, ChmCGG, and CghmCGG. Therefore, T4-β-GT combined with Msp I/Hpa II digestion assay was conducted to detect ChmCGG and CmCGG, especially the unsymmetrical hydroxymethylation (30-32). Briefly, the glucosylation reactions lasted for 16 h at 37 °C, then the glucosylation was divided into four aliquots for different subsequent treatments (Msp I treatment, no Msp I control, Hpa II treatment, and no Hpa II control).
Then, 50 units of Msp I or Hpa II (New England Biolabs) were added into the corresponding aliquots; and no restriction enzyme was added into the controls. After incubated for 10 h at 37 °C, 1 µL proteinase K (20 mg/mL) was added to the digested glucosylation and incubated at 40 °C for 30 min, and then incubated at 95 °C for 10 min. The CCGG-containing CDKN2A promoter fragment was amplified by end point PCR with primer set 5’-gggctctcac aactaggaa-3’ and 5’-cggaggaggt gctattaactc-3’, as previously reported (10). The CCND1 gene containing an unmethylated-KBS sequence was used as the positive ChIP-PCR control, amplified with the primer set (sense 5’-tttacatctg cttaagtttg cg-3’ and antisense 5’-ttagaatttg ccctgggact-3’). The 166 bp GAPDH fragment containing an unmethylated-CCGG site was used as the negative ChIP-PCR control, amplified with the primers (sense, 5’-tactagcgtt ttacgggcg-3’; antisense, 5’-tcgaacagga ggagcagaga gcga-3’).
Results
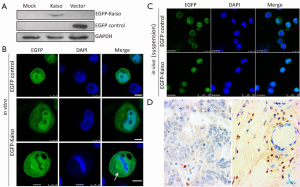
Most of Kaiso locates within the nucleus in vivo
To track subcellular localization of Kaiso, the EGFP-Kaiso expression vector was stably transfected into MGC803 cells. Results of Western blotting confirmed the expression of EGFP-Kaiso in the transfected cells (Figure 1A). EGFP-fluorescence examined by confocal microscope showed that Kaiso existed both in the nucleus and the cytoplasm (Figure 1B). In addition, the co-localization of Kaiso with the spindle microtubules was found in cells at metaphase (Figure 1B, bottom), which was consistent with previous reports (22,23), indicating distinctive visibility of the transfected Kaiso in cells. Next, MGC803 cells, with EGFP-Kaiso or EGFP control stable transfection, were inoculated into the peritoneum of nude mice. While the EGFP control was mainly localized in the cytoplasm surrounding the nucleus, most of EGFP-Kaiso was observed in the nucleus of cells in the peritoneum fluid (Figure 1C), suggesting a shift of the cytoplasmic Kaiso to the nucleus in vivo. IHC analysis also showed the endogenous Kaiso protein was mainly and clearly located in nucleus of stromal cells in the representative primary gastric carcinoma tissues, cytoplasm Kaiso staining, if any, was weak (Figure 1D).
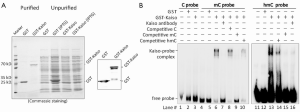
Kaiso is able to bind with methylated- and hydroxymethylated-CGCG sequences in vitro
Then we investigated the exact DNA binding sequences of Kaiso. Kaiso’s accessibilities to unmethylated-, methylated-, and hydroxymethylated-CGCG containing CDKN2A sequences were analyzed in vitro. GST-Kaiso was induced and purified from E. coli (Figure 2A). Results of EMSA analysis showed that the purified GST-Kaiso was able to bind to both hydroxymethylated- and methylated-CDKN2A probes (Figure 2B lane #6 and #13), but not with the unmethylated-CDKN2A probe (lane #3). In the competitive assay, Kaiso’s binding with the hydroxymethylated-probe was partially inhibited by 10 times of the unlabeled hydroxymethylated-probe (lane #14), but completely inhibited by 10 times of the methylated probe and Kaiso-antibody (lane #15 and #16), indicating that Kaiso’s binding affinity to the hydroxymethylated-DNA is weaker than to the methylated-one in vitro.
Kaiso is not able to bind with hydroxymethylated-CGCG sequence in vivo
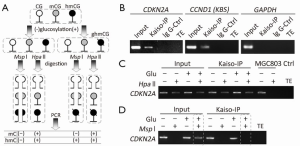
Recently, we have reported that the CpG islands surrounding transcription start site of some wild-type CDKN2A alleles are actually fully hydroxymethylated in HCT116 cells (a CDKN2A hemi-methylated cell line) (33). In order to confirm the results observed in the above in vitro analysis, we further determined whether Kaiso binds with the methylated- and hydroxymethylated-CDKN2A promoter in HCT116 cells using ChIP assay combined with CCGG-specific restriction enzymes Hpa II and Msp I (Figure 3A). As expected, the CDKN2A promoter was efficiently amplified from the chromatin immunoprecipitated by the Kaiso-antibody (Kaiso-IPed chromatin), but not amplified from the chromatin immunoprecipitated by the IgG control antibody (Figure 3B, the left panel). Similarly, the KBS-containing CCND1 was also successfully amplified (Figure 3B, the middle panel). In addition, the GAPDH promoter control, containing an unmethylated-CGCG, was not amplified (Figure 3B, the right panel). Overall, these results are consistent with previous reports that Kaiso specifically bind with the methylated-CGCG and unmethylated-KBS sequences in vivo.
We further treated the Kaiso-IPed chromatin by T4-β-GT glucosylation combined with or without Msp I/Hpa II restriction digestion. Both Msp I and Hpa II digest the unmethylated CCGG sites. Msp I also digests methylated- and hydroxymethylated-CCGGs (CmCGG and ChmCGG), but cannot digest glucosylated-ChmCGG (CghmCGG); while Hpa II cannot digest any modified CCGG such as CmCGG, ChmCGG and CghmCGG. Therefore, T4-β-GT combined with Msp I/Hpa II digestion assay was conducted to detect ChmCGG and CmCGG, especially the unsymmetrical hydroxymethylation (29,30,32). As shown in Figure 3C, the CDKN2A promoter was amplified in the Kaiso-IPed and Hpa II-digested chromatin from HCT116 cells, but was not amplified in the Hpa II-digested input control chromatin from MGC803 cells (a CDKN2A-unmethylated cell line), indicating specific binding of Kaiso with the methylated (or hydroxymethylated, if any) CDKN2A alleles. Following the glucosylation treatment and Msp I-digestion, the CDKN2A promoter can be still amplified from the input chromatin (Figure 3D), consistent with our report on hydroxymethylation of some CDKN2A CpG islands in HCT116 cells (33). However, the CDKN2A promoter was not amplified from the Kaiso-IPed chromatin, implying no Kaiso binding to the hydroxymethylated-CDKN2A alleles. Together, the ChIP-glucosylation-restriction-PCR analysis showed that Kaiso could bind with the methylated-CDKN2A, but not with hydroxymethylated-CDKN2A in vivo.
Discussion
It has been reported that intracellular location of Kaiso is different in cancer cells under different growth condition (21). In the present study, the translocation phenomenon of Kaiso was confirmed using direct EGFP tracking tag. We found that Kaiso was located both in the nucleus and cytoplasm of cultured cells in vitro, but mainly located in the nucleus in human gastric cancer MGC803 cells in the peritoneum fluid in nude mice and stromal cells in primary gastric carcinoma tissues. Most importantly, to our knowledge, we first report that nuclear Kaiso could bind with the methylated-CGCG, but could not bind with hydroxymethylated-CGCG sequences in vivo.
In previous research, nuclear Kaiso was rarely found in tissues by means of immuno-detection. This challenges whether Kaiso actually engages in transcriptional repression in vivo. In our study, both endogenous and EGFP-tag Kaiso existed predominantly in the nucleus, providing strong evidences to support Kaiso as a transcriptional repressor. Although we have confirmed the shift of Kaiso between nucleus and cytoplasm, the underlying mechanism of translocation is still unknown. It has been reported that Kaiso might shuttle out of the nucleus via phosphorylated p120ctn isoform 3 (25). Further studies are needed to address sequence-specific binding of Kaiso to its target genes.
CDKN2A is a tumor suppressor gene that is frequently inactivated by DNA methylation in cancer and precancerous lesions (34). It has been previously reported that Kaiso binds with the methylated-CDKN2A in HCT116, HT29 M6, and SW480 cell lines, but not with the unmethylated-CDKN2A in HEK293T cell line (10,35). Here we found similar interactions between Kaiso and the CDKN2A promoter in HCT116 cells by Kaiso-IPed-chromatin analysis. Recently, we have found that some CpG islands in the wild-type CDKN2A alleles are completely hydroxymethylated in HCT116 cells using the hMeDIP-PCR and TAB-seq assays (33). In addition, β-GT-glucosylation combined with Msp I-restriction and subsequent PCR is a very sensitive assay to detect hydroxymethylation at the CCGG sites (30). Using the combined assay, hydroxymethylated-CDKN2A was detected in HCT116 cells once again. However, the interaction between the nuclear Kaiso and CDKN2A was not observed in HCT116 cells, though this interaction was observed in the EMSA analysis.
Kinetic analysis has revealed that Msp I activity is dramatically decreased by symmetrical hydroxymethylation of its recognition sequence and partly inhibited by hemi-hydroxymethylation (31). CDKN2A is asymmetrically hydroxymethylated in HCT116 cells (33). The fact that no CDKN2A PCR products could be amplified indicates that all of the CDKN2A templates in the Kaiso-IPed chromatin were completely digested. Therefore, the possibility of interaction between the nuclear Kaiso and the hydroxymethylated CDKN2A alleles could be excluded.
Conclusions
The present study confirms that nuclear Kaiso only binds to methylated-CGCG, but not hydroxymethylated-CGCG sequences in the genome in vivo. Kaiso cannot be a binding protein for hydroxymethylated DNA.
Acknowledgements
This study was supported by National Basic Research Program of China (Grant No. 2011CB504201). We appreciate Drs. Shengyan Xiang and Kendra Allana Williams for their English language editing.
Disclosure: The authors declare no conflict of interest.
References
- Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol 1999;19:3614-23. [PubMed]
- Daniel JM, Spring CM, Crawford HC, et al. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res 2002;30:2911-9. [PubMed]
- Prokhortchouk A, Hendrich B, Jørgensen H, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev 2001;15:1613-8. [PubMed]
- Zhang BZ, Gu LK, Deng DJ. Methylation specific binding activity of zinc finger protein Kaiso. Zhonghua Yu Fang Yi Xue Za Zhi 2007;41:43-6. [PubMed]
- Blattler A, Yao L, Wang Y, et al. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin 2013;6:13. [PubMed]
- Kim SW, Fang X, Ji H, et al. Isolation and characterization of XKaiso, a transcriptional repressor that associates with the catenin Xp120(ctn) in Xenopus laevis. J Biol Chem 2002;277:8202-8. [PubMed]
- Park JI, Kim SW, Lyons JP, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell 2005;8:843-54. [PubMed]
- Spring CM, Kelly KF, O’Kelly I, et al. The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp Cell Res 2005;305:253-65. [PubMed]
- Prokhortchouk A, Sansom O, Selfridge J, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol 2006;26:199-208. [PubMed]
- Lopes EC, Valls E, Figueroa ME, et al. Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res 2008;68:7258-63. [PubMed]
- Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011;473:398-402. [PubMed]
- Gu TP, Guo F, Yang H, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011;477:606-10. [PubMed]
- Ruzov A, Tsenkina Y, Serio A, et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res 2011;21:1332-42. [PubMed]
- Dickson KM, Gustafson CB, Young JI, et al. Ascorbate-induced generation of 5-hydroxymethylcytosine is unaffected by varying levels of iron and 2-oxoglutarate. Biochem Biophys Res Commun 2013;439:522-7. [PubMed]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 2010;38:e125. [PubMed]
- Yildirim O, Li R, Hung JH, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 2011;147:1498-510. [PubMed]
- Hashimoto H, Liu Y, Upadhyay AK, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 2012;40:4841-9. [PubMed]
- Mellén M, Ayata P, Dewell S, et al. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012;151:1417-30. [PubMed]
- Baubec T, Ivánek R, Lienert F, et al. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell 2013;153:480-92. [PubMed]
- Zhenilo SV, Musharova OS, Pokhorchuk EB. Transcription factor Kaiso does not interact with hydroxymethylated DNA within CTGCNA sequence context. Mol Biol (Mosk) 2013;47:522-5. [PubMed]
- Soubry A, van Hengel J, Parthoens E, et al. Expression and nuclear location of the transcriptional repressor Kaiso is regulated by the tumor microenvironment. Cancer Res 2005;65:2224-33. [PubMed]
- Kantidze OL, Kamalyukova IM, Razin SV. Association of the mammalian transcriptional regulator kaiso with centrosomes and the midbody. Cell Cycle 2009;8:2303-4. [PubMed]
- Soubry A, Staes K, Parthoens E, et al. The transcriptional repressor Kaiso localizes at the mitotic spindle and is a constituent of the pericentriolar material. PLoS One 2010;5:e9203. [PubMed]
- Kelly KF, Otchere AA, Graham M, et al. Nuclear import of the BTB/POZ transcriptional regulator Kaiso. J Cell Sci 2004;117:6143-52. [PubMed]
- Zhang PX, Wang Y, Liu Y, et al. p120-catenin isoform 3 regulates subcellular localization of Kaiso and promotes invasion in lung cancer cells via a phosphorylation-dependent mechanism. Int J Oncol 2011;38:1625-35. [PubMed]
- Dai SD, Wang Y, Jiang GY, et al. Kaiso is expressed in lung cancer: its expression and localization is affected by p120ctn. Lung Cancer 2010;67:205-15. [PubMed]
- Dai SD, Wang Y, Miao Y, et al. Cytoplasmic Kaiso is associated with poor prognosis in non-small cell lung cancer. BMC Cancer 2009;9:178. [PubMed]
- Wang KH. An in vitro cell line (MGc80-3) of a poorly differentiated mucoid adenocarcinoma of human stomach. Shi Yan Sheng Wu Xue Bao 1983;16:257-67.
- Donaldson NS, Pierre CC, Anstey MI, et al. Kaiso represses the cell cycle gene cyclin D1 via sequence-specific and methyl-CpG-dependent mechanisms. PLoS One 2012;7:e50398. [PubMed]
- Davis T, Vaisvila R. High sensitivity 5-hydroxymethylcytosine detection in Balb/C brain tissue. J Vis Exp 2011;2661. [PubMed]
- Ichiyanagi K. Inhibition of MspI cleavage activity by hydroxymethylation of the CpG site: a concern for DNA modification studies using restriction endonucleases. Epigenetics 2012;7:131-6. [PubMed]
- Bhattacharyya S, Yu Y, Suzuki M, et al. Genome-wide hydroxymethylation tested using the HELP-GT assay shows redistribution in cancer. Nucleic Acids Res 2013;41:e157. [PubMed]
- Qin S, Li Q, Zhou J, et al. Homeostatic maintenance of allele-specific p16 methylation in cancer cells accompanied by dynamic focal methylation and hydroxymethylation. PLoS One 2014;9:e97785. [PubMed]
- Deng DJ, Li Q, Wang XH. Methylation and demethylation of Ink4 locus in cancer development. Chin J Cancer Res 2010;22:245-52.
- Del Valle-Pérez B, Casagolda D, Lugilde E, et al. Wnt controls the transcriptional activity of Kaiso through CK1ε-dependent phosphorylation of p120-catenin. J Cell Sci 2011;124:2298-309. [PubMed]


