microRNA-218 suppresses the proliferation, invasion and promotes apoptosis of pancreatic cancer cells by targeting HMGB1
Introduction
Pancreatic cancer is a malignant tumor of the digestive system featured by insidious clinical manifestation, difficult early diagnosis, rapid progression, and extremely poor prognosis (1-3). With worldwide increase in prevalence, pancreatic cancer has become the fourth leading cause of cancer-related death. Most pancreatic cancer patients have already in the advanced stages when the disease is confirmed, and their 5-year survival rate is less than 5% (4,5). Epidemiological data have shown that a higher incidence of pancreatic cancer is associated with factors including smoking, high-fat high-protein diets, alcoholism, coffee drinking, exposure to certain chemical carcinogens, diabetes mellitus, and chronic pancreatitis (6-8). Therefore, it is critically important to carry out basic research on the pancreatic cancer, so as to identify are highly sensitive and specific novel tumor markers (especially feasible for the early diagnosis) and develop new diagnosis and treatment techniques (9-11). Along with the development of the theories and techniques of molecular biology, new advances have been continuously made in the pancreatic cancer-related genes and new hot research topics have emerged (12,13).
According to the basic theories of molecular biology, oncogenesis is a multi-step progress involving the activation of multiple cancer genes and the inactivation of tumor suppressor genes, which cause the abnormal growth of cells as well as the changes in phenotypes and functions; as a result, the normal cells may be transformed into tumor cells (14-16). After many years of efforts, many achievements have been made in terms of oncogenes, tumor suppressor genes, and signal transduction pathways; however, the exact mechanism governing the development of pancreatic cancer remains unclear (17,18).
MicroRNAs (miRNAs) are endogenous, single-stranded, small, ~22-nucleotide noncoding RNAs. miRNAs generally rregulate the mRNA expression at the post-transcriptional level by base pairing to complementary sequences within the 3’ untranslated region (3’UTR) of protein-coding mRNA transcripts, and thus are involved in various physiological processes including the proliferation, apoptosis, differentiation, metabolism, and development of cells and pathological conditions such as cardiovascular diseases, neurological diseases, and tumors (19-21). It is estimated that more than one third of human genes are regulated by miRNAs. The roles of miRNA in human diseases, in particular in tumors, are increasingly concerned (22,23).
Recent studies have demonstrated that miRNA is invovled in the occurence, development, and invasion of tumors by regulating the expressions of the genes that are related with the cell cycle, apoptosis, cell migration, and angiogenesis (24-26). miRNAs are found to be closely associated with the development of tumors. Similar studies have also been carried out in pancreatic cancer (27,28). However, relatively few studies have been carried out on the role of miRNA in pancreatic cancer (29). In their study on the brain gliomas, Tu et al. found that miR-218 could suppress the proliferation and migration of glioma cells by regulating the target gene Bmi1 (30). A study on the osteogenic sarcoma showed that miR-218 could also suppress the invasion and migration of osteosarcoma cells by regulating the targeting genes including TIAM1, MMP2, and MMP9 (31). Zhu et al. demonstrated that miR-218 was expressed in the pancreatic ductal adenocarcinima (PDAC) and was associated with the development of pancreatic cancer (32). While miR-218 may be a new marker of pancreatic cancer, the effects of miR-218 on the biological characteristics of pancreatic cancer cells as well as the underlying mechanisms remain unclear.
In our current study, we tried to determine the expression profiles of miR-218 in human pancreatic cancer tissue (PCT) and cells and their effects on the biological features of human pancreatic cancer cell line PANC-1 and observe the impact of miR-218 on the expression of the target gene HMGB1, with an attempt to provide new treatment methods and strategies for pancreatic cancer.
Materials and methods
Specimen
Fresh tumor specimens were surgically isolated from ten patients with pancreatic carcinoma who were treated in our hospital. Also, normal pancreas tissues were harvested from five subjects. All these specimens were immediately preserved in liquid nitrogen until subsequent experiments. All patients were naive to any radiotherapy, chemotherapy, or other therapies and had no other inflammatory disease.
Main reagents
Fetal calf serum, Dulbecco’s modified Eagle’s medium (DMEM), RPMI 1640 medium, L-glutamine, HEPES, and Lipofectamine 2000 were purchased from Invitrogen, USA.
TaqMan miRNA isolation kit, TaqMan microRNA assay kit, TaqMan microRNA assay, and TaqMan Universal PCR Master Mix were purchased from Applied Biosystems. The miR-218 RT primers, miR-218 mimic, and non-specific control (mimic ctrl) were synthesized by Shanghai Genepharma Corporation.
Cell Counting Kit-8 (CCK-8) was purchased from Beyotime Institute of Biotechnology (Shanghai, China). The trypsin, phosphate buffered saline (PBS), and Hoechst 33342 were purchased from Sigma-Aldrich (USA). Cell culture plates or dishes and Transwell invasion chamber were purchased from Corning, USA. Matrigel gum was purchased from BD Bioscience. Annexin V and propidium iodide (PI) were purchased from Roche, Germany. The primary antibodies including rabbit anti-human HMGB1 monoclonal antibody and mouse anti-human β-actin monoclonal antibody were purchased from Abcam, UK. The secondary antibodies including HRP-conjugated affinity purified goat anti-mouse IgG and HRP-conjugated affinity purified goat anti-rabbit IgG were purchased from Sigma-Aldrich. Protein extraction and quantification kit was purchased from Bio-Rad company.
Methods
Cell culture and treatment
The AsPC-1 and BxPC-3 cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 10 mM HEPES, 1.5 g/L NaHCO3, and 2 mM L-glutamine. The PANC-1 cells were cultured in DMEM containing 10% FBS, 1.5 g/L NaHCO3, and 4 mM L-glutamine. All the AsPC-1, BxPC-3, and PANC-1 cells were inoculated at 37 °C, 5% CO2 and saturated humidity. The growth of cells was observed under an inverted microscope. When cells were grown to 70-80% confluence, they were detached with 0.25% trypsin. Medium was changed every other day, and the cells were passaged every 4 to 6 days. Cells in the exponential phase were selected for further experiment.
The normally cultured PANC-1 cells were uniformly seeded in 6-well culture plates at a density of 3×105/mL (1,000 µL in each well). After the cells reached complete adherence, the transfections of miR-218 mimic and non-specific control (mimic ctrl) were performed using Lipofectamine 2000 in accordance with the manufacturer’s instruction. Meanwhile, normal control (normal ctrl) group was set. Serum-free minimum essential medium (MEM) was used to dilute the miR-218 mimic and mimic ctrl. Then, the liposome Lipofectamine 2000 was diluted in the MEM, gently mixed, and then inoculated at room temperature for 5 min. The diluted Lipofectamine 2000 was separately mixed with the diluted miR-218 mimic and mimic ctrl and then inoculated at room temperature for 20 min to form complexes. The complexes were then added into the culture plate containing the PANC-1 cells, which was put inside an incubator at 37 °C in the humidified incubator with 5% CO2. Five hours later, MEM containing 10% FBS was used instead, and the cells were cultured for another 48 h.
Detection of the miR-218 expression in human PCT and cell lines
The expressions of miR-218 in pancreatic cancers including AsPC-1, BxPC-3, and PANC-1 and in PCT and normal pancreatic tissue (NPT) were determined using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). After the in vitro cultured pancreatic cancer cells and tissues were collected, the RNA was extracted using the TaqMan miRNA isolation kit, the expression of the mature miR-218 was determined using the TaqMan microRNA assay and TaqMan Universal PCR Master Mix, with U6 being an internal control. Three duplicate wells were set in each group for the assay. The CT value of the specimen in each reaction tube was recorded. The relative quantification method in the qRT-PCR was employed for analyzing the experimental results.
Detection of the effect of the transfection of miR-218 mimic in PANC-1 cells on the expression of miR-218
Forty-eight hours after the transfection of miR-218 mimic or mimic ctrl in the PANC-1 cells, the RNA of cells in each group was extracted using the TaqMan miRNA isolation kit and the changes in the miR-218 expression in the PANC-1 cells in each group were determined using the qRT-PCR.
Detection of the effect of miR-218 on cell viability using the CCK-8 method
Based on the manufacturer’s instructions, the CCK-8 solution was added in each well 1, 2, 3, and 4 days after the transfection of miR-218 mimic or mimic ctrl in the PANC-1 cells, and then the cells were incubated at 37 °C in a 5% CO2 incubator for 2 hours. Meanwhile, normal control (normal ctrl) group was set. The OD value was measured at 450 nm using a microplate reader (Bio-Tek, USA), using a reference wavelength of 630 nm. Ten duplicate wells were used for each experimental group, and the experiment was repeated three times.
Determination of the effect of miR-218 on cell proliferation using flow cytometry
The PANC-1 cells were transfected with miR-218 mimicor mimic ctrl. Meanwhile, normal control (normal ctrl) group was set. Wash the cells 1-2 times with PBS 48 h after the transfection. Wash the cells twice with PBS after the cells were detached with trypsin. Cells were collected by centrifugation, and then added with PI staining solution and incubated 30 min in the dark at 4 °C. After the mesh filter, the apoptosis was detected using flow cytometry (BD Company, USA). The cells were counted using the CellQuest software (BD, USA), and the data were analyzed using the Macquit software.
Determination of the effect of miR-218 on cell apoptosis using flow cytometry
The PANC-1 cells were transfected with miR-218 mimicor mimic ctrl. Meanwhile, normal control (normal ctrl) group was set. Wash the cells 1-2 times with PBS 48 h after the transfection. Adding Annexin V-FITC and PI dye, and then let incubate at room temperature in dark for 15 min. After the mesh filter, the apoptosis was detected using flow cytometry (BD Company, USA). The cells were counted using the CellQuest software (BD, USA), and the data were analyzed using the Macquit software.
Determination of the effect of miR-218 on cell migration using the Trans-well migration assay
The transfections of miR-218 mimic and non-specific control (mimic ctrl) were performed using Lipofectamine 2000 in accordance with the manufacturer’s instructions. Meanwhile, normal control (normal ctrl) group was set. Twenty-four hours after the transfection, the PANC-1 cells were inoculated in Transwell invasion chamber. The cells were cultured in the normal conditions for 24 hours, then washed with PBS 1-2 times and stained with Hoechst 33342. Count the number of cells that had passed through the Transwell polycarbonate membrane using Leica microscope (Leica company, Wetzlar, Germany). These cells were regarded as invasive cells, and eight random fields were observed.
Determining whether HMGB1 was the target gene of miR-218 by using the luciferase reporter assay
Construction of the luciferase reporter plasmid: After the sequence fragments of the target gene HMGB1 3’-UTR complementary to miR-218 and the mutated HMGB1 3’-UTR sequence fragments were synthesized using chemical approaches, the HMGB1 3’-UTR (wild type, WT) that was used for predicting miR-218 binding sites and the mutated HMGB1 3’-UTR (mutant) were cloned into the Xba I site of the pGL3-Lueiferase vector.
The normally cultured HEK293 cells were uniformly seeded in 6-well culture plates at a density of 3×105/mL (1,000 µL in each well). The luciferase plasmid containing the HMGB1 3’-UTR(WT) and the mutated HMGB1 3’-UTR (mutant) as well as miR-218 mimic or mimic ctrl were transfected. Meanwhile, the normal control (normal ctrl) group was set. Cells were collected 48 hours after the transfection and detected using the luciferase kit, and the results were read using an ELISA reader.
The effect of miR-218 on the HMGB1 protein expression was determined using Western blotting. The cellular proteins in each group were collected 48 h after the transfection of miR-218 mimic or mimic ctrl in PANC-1 cells. These cells placed in 6-well cell culture plates. Add 1 mL of RIPA lysis buffer [150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 7.9), 10 mM NaF, 10 mM PMSF and 1× protease inhibitors (Complete cocktail tablets, Roche)] in each well. Then, the cell lysates were transferred to a 1.5 mL centrifuge tube and centrifuged at 16,000 rpm for 30 min. The protein concentration of the supernatant was determined using bicinchoninic acid (BCA) method. Electrophoretic separation was conducted using 5% stacking gel and 15% separating gel, with 50 µg total protein added in each lane. After the protein had been wet transferred onto a PVDF membrane (Bio-Rad, USA), they were blocked in TBST solution (10 mM Tris-HCl, pH7.5, 150 mM NaCl, and 0.1% Tween-20) containing 5% skimmed milk powder at room temperature for 1 h, then added with rabbit anti-human HMGB1 antibody (1:500 dilution) and rabbit anti-human β-actin multiclonal antibody (1:1,000 dilution) and incubated overnight at 4 °C. Wash three times with fresh TBST for 5 min. Add HRP-labeled goat anti-rabbit IgG or HRP-labeled goat anti-mouse IgG secondary antibody and inoculate it at 37 °C for 1 h. Wash three times with fresh TBST for 5 min. Autoradiography was conducted using ECL chemiluminescent substrate. The relative concentration of HMGB1 was presented as RKIP/β-actin ratio, and the change of its relative expression was analyzed using PDQuest software (Bio-Rad, Richmond, CA, USA).
Statistical analysis
The experimental data were analyzed using SPSS 17.0 software. Inter-group differences were analyzed by t-test and multiple comparisons among several groups were conducted by analysis of variance. P<0.05 was considered significantly different.
Results
The miR-218 expressions in human PCT and cell lines
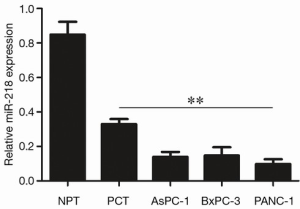
As shown by qRT-PCR, the expression of miR-218 in PCT was significantly lower than in NPT (P<0.01), and its expression in pancreatic cancer cell lines (AsPC-1, BxPC-1, and PANC-1) were significantly lower than those in the NPT (P<0.01) (Figure 1).
The effect of the transfection of miR-218 mimic in PANC-1 cells on the expression of miR-218
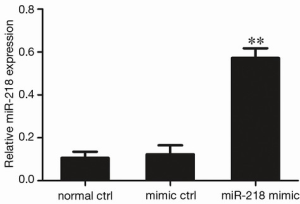
The miR-218 was over-expressed in PANC-1 cells by the transfection of miR-218 mimic in these cells. As shown by qRT-PCR, the MiR-218 expression in the miR-218 mimic transfection group was significantly higher than that in the normal control (normal ctrl) group (P<0.01) and negative control (mimic ctrl) group (P<0.01) (Figure 2).
Detection of the effect of miR-218 on cell viability using the CCK-8 method
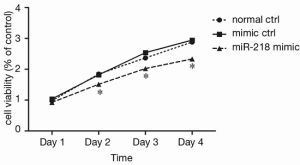
As shown by CCK-8 analysis, 2 days after the miR-218 mimic transfection, the cell viability in the miR-218 mimic transfection group was significantly lower than that in the normal control (normal ctrl) group (P<0.05) and negative control (mimic ctrl) group (P<0.05) (Figure 3).
The effect of miR-218 on cell proliferation using flow cytometry
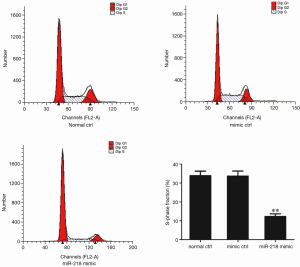
The cell proliferation in each group was detected using flow cytometry 48 h after the transfection of miR-218 mimic or mimic ctrl in PANC-1 cells. S-phase fraction = (S)/(G0G1+S+G2M). As shown by flow cytometry, the S-phase fraction in the miR-218 mimic transfection group was significantly lower than that in the normal control (normal ctrl) group (P<0.01) and negative control (mimic ctrl) group (P<0.01) (Figure 4).
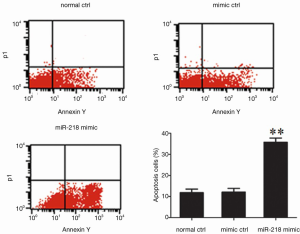
Determination of the effect of miR-218 on cell apoptosis using flow cytometry
The S-phase fraction in the miR-218 mimic transfection group was significantly lower than that in the normal control (normal ctrl) group (P<0.01) and negative control (mimic ctrl) group (P<0.01) (Figure 5).
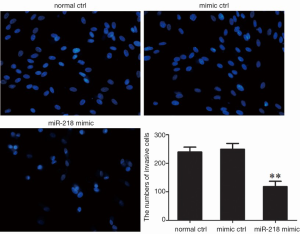
The effect of miR-218 on cell migration using the Trans-well migration assay
The cell migration capacity in each group was detected using Trans-well migration assay 48 h after the transfection of miR-218 mimic or mimic ctrl in PANC-1 cells. As shown by Hoechst 33342 staining, the transfection with miR-218 mimic was associated with a significantly lower number of cells that passed through a Transwell chamber (P<0.01) (Figure 6).
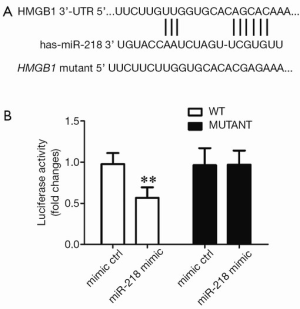
Determining whether HMGB1 was the target gene of miR-218 by using the luciferase reporter assay
To validate whether the putative target site of miR-218 is located atHMGB1 3’-UTR, we constructed a luciferase plasmid containing theHMGB1 3’-UTR (WT) and the mutated HMGB1 3’-UTR (mutant) and used it to co-transfect the HEK293 cells with miR-218 mimic or mimic ctrl. Compared with the mimic ctrl group, the luciferase activity in the plasmid significantly decreased in the miR-218 mimic transfection group (P<0.01) (Figure 7).
The regulatory effect of miR-218 on HMGB1 expression was detected by using Western blotting, which showed that the HMGB1 protein expression in the miR-218 mimic transfection group was significantly lower than those in the normal control (normal ctrl) group (P<0.01) and negative control (mimic ctrl) group (P<0.01) (Figure 8).

Discussion
Pancreatic cancer is one of the most common forms of gastrointestinal malignancies. As a highly malignant tumor, its prognosis is extremely poor due to its deep anatomic position, insidious onset, difficulty in early diagnosis, poor response to radiochemotherapy, and inoperability (33,34). Therefore, in-depth research on the occurrence and development of pancreatic cancer is urgently required to improve its early diagnosis and optimize the treatment (35,36).
Similar to many other malignancies, the pathogenesis of pancreatic cancer is also a progressive process involving multiple genes, steps, and stages. Therefore, gene therapy has been recognized as one of the most promising strategies (9,37,38). Numerous studies have shown that miRNA disturbances appear during the tumorigenesis (39,40). When over-expressed in tumor cells, some miRNAs (e.g., miR-21 and miR-10b) can induce tumor invasion/metastasis and thus exert the oncogene-like functions. These miRNAs are known as carcinogenic miRNAs (41). On the other hand, the down-regulation of some other miRNAs (e.g., miR-335, miR146a, and miR-29c) is associated with the occurence, development, and invasion/metastasis of tumors. They are called as tumor suppressor miRNAs (42). As shown by a previous study, miR-218 is regarded as a tumor suppressor miRNA can is down-regulated in many tumors (43). Shi et al. found that the expression of miR-218 was down-regulated in childhood medulloblastoma (44). Tu et al. reported that miR-218 was down-regulated in glioma and could suppress the proliferation, migration, and invasion of glioma cells (30). In our current study, by using qRT-PCR, we detected the expressions of miRNA-218 in PCT, NPT, and pancreatic cancer cell lines. As shown by qRT-PCR, the expression of miR-218 in PCT was significantly lower than in NPT, and its expressions in pancreatic cancer cell lines (AsPC-1, BxPC-1, and PANC-1) were significantly lower than that in the NPT. Therefore, miR-218 may also act as a tumor suppressor miRNA in pancreatic cancer.
According to Guan et al., the expression of miR-218 was down-regulated in thyroid cancer, and the down-regulated miR-218 was associated with the invasion of thyroid cancer cells and the progression of thyroid cancer (45). In our current study, we used CCK-8, flow cytometry, and Transwell assay to explore the effects of miR-218 on the biological characteristics of pancreatic cancer cell line PANC-1. As shown in our study, after the over-expression of miR-218 in PANC-1 cells, the viability of PANC-1 cells decreased, the proliferation and invasion of PANC-1 cells were suppressed, and the apoptosis was promoted. Therefore, our study demonstrated that miR-218 also served as a tumor suppressor miRNA in PANC-1 cells.
HMGB1 is a member of the high mobility group box family of non-histone chromosomal proteins (46-48). Studies have shown that, after binging with DNA, HMGB1 is involved in the construction and maintenance of the nucleosome and thus plays different roles in the transcription, replication, recombination, DNA repair, and maintenance of genome stability (49,50). In addition, HMGB1 can acts as an inflammatory factor after having been actively secreted by inflammatory cells and (or) tumor cells or passively secreted from necrotic cells into the extracellular environment, in which it binds to multiple receptors [mainly the receptor for advanced glycation end product (RAGE)] on cell surface and activates a series of downstream signaling pathways; thus, it plays vital roles in inflammation, cancer, sepsis, autoimmune disease, and vascular diseases (51,52). In our current study, the online software TARGETSCAN was employed to analyze and predict that HMGB1 was a target gene of miR-218. We also validated that the putative target site of miR-218 is located at HMGB1 3’-UTR, as shown by the luciferase reporter assay. Also, the regulatory effect of miR-218 on HMGB1 expression was detected by using Western blotting, which showed that the HMGB1 protein expression in the miR-218 mimic transfection group was significantly lower than those in the normal control (normal ctrl) group and negative control (mimic ctrl) group. Thus, the miR-218 overexpression inhibited the HMGB1 protein expression.
Stimulated by cytokines such as growth factors, tumor cells can excessively express and secret HMGB1 (46,53,54). By binding to its receptor RAGE, the secretory HMGB1 can stimulate the proliferation, migration, and invasion of cells and the angiogenesis, and thus promote tumorigenesis (55,56). Meng et al. demonstrated that the overexpression of HMGB1 in osteogenic sarcoma cell line MG-63 could promote the proliferation and invasion of MG-3 cells and inhibit apoptosis (56). Wang et al. used the lentivirus-mediated RNA to intervene and inhibit the expression of HMGB1 in prostate cancer cells and found that the suppression of HMGB1 gene expression inhibited the proliferation of prostate cancer cells and promoted their apoptosis (57). In our current study, we firstly confirmed that HMGB1 was the target gene of miR-218 and then demonstrated that miR-218 inhibited the HMGB1 protein expression. Thus, miR-218 may inhibit the proliferation and invasion of pancreatic cancer cell line PANC-1 and promote its apoptosis by suppressing the expression of its target gene HMGB1.
In conclusion, the miR-218 expression decreases in human PCT and cell lines. miR-218 can negatively regulate the HMGB1 protein expression and inhibit the proliferation and invasion of pancreatic cancer cells. A treatment strategy by enhancing the miR-218 expression may benefit the patients with pancreatic cancer.
Acknowledgements
Funding: This study was supported by grants: (I) Liaoning Provincial Department of education science research project (L2014299); (II) Liaoning province science and technology plan project (2011404013-4); (III) The Shenyang Municipal Science and technology project (F12-277-1-73).
Disclosure: The authors declare no conflict of interest.
References
- Chen W, Zheng R, Zhang S, et al. Annual report on status of cancer in China, 2010. Chin J Cancer Res 2014;26:48-58. [PubMed]
- Rossi RE, Massironi S, Conte D, et al. Therapy for metastatic pancreatic neuroendocrine tumors. Ann Transl Med 2014;2:8. [PubMed]
- Arslan C, Yalcin S. Current and future systemic treatment options in metastatic pancreatic cancer. J Gastrointest Oncol 2014;5:280-95. [PubMed]
- Rozengurt E. Mechanistic target of rapamycin (mTOR): a point of convergence in the action of insulin/IGF-1 and G protein-coupled receptor agonists in pancreatic cancer cells. Front Physiol 2014;5:357. [PubMed]
- Liu J, Song R, Cui M. Numerical simulation on hydromechanical coupling in porous media adopting three-dimensional pore-scale model. ScientificWorldJournal 2014;2014:140206.
- Moir J, White SA, French JJ, et al. Systematic review of irreversible electroporation in the treatment of advanced pancreatic cancer. Eur J Surg Oncol 2014;40:1598-604. [PubMed]
- Burkey MD, Feirman S, Wang H, et al. The association between smokeless tobacco use and pancreatic adenocarcinoma: a systematic review. Cancer Epidemiol 2014;38:647-53. [PubMed]
- Ito M, Makino N, Ueno Y. Glucose intolerance and the risk of pancreatic cancer. Transl Gastrointest Cancer 2013;2:223-29.
- Furuse J, Nagashima F. Current status and future direction of chemotherapy for pancreatic cancer. Chin Clin Oncol 2013;2:6. [PubMed]
- Xu K, Niu L, Yang D. Cryosurgery for pancreatic cancer. Gland Surg 2013;2:30-9. [PubMed]
- Garajová I, Le Large TY, Frampton AE, et al. Molecular mechanisms underlying the role of microRNAs in the chemoresistance of pancreatic cancer. Biomed Res Int 2014;2014:678401.
- Valle JW, Faivre S, Hubner RA, et al. Practical management of sunitinib toxicities in the treatment of pancreatic neuroendocrine tumors. Cancer Treat Rev 2014;40:1230-8. [PubMed]
- Chitkara D, Mittal A, Mahato RI. miRNAs in pancreatic cancer: therapeutic potential, delivery challenges and strategies. Adv Drug Deliv Rev 2015;81:34-52. [PubMed]
- Pulito C, Donzelli S, Muti P, et al. microRNAs and cancer metabolism reprogramming: the paradigm of metformin. Ann Transl Med 2014;2:58. [PubMed]
- Chen BJ, Wu YL, Tanaka Y, et al. Small molecules targeting c-Myc oncogene: promising anti-cancer therapeutics. Int J Biol Sci 2014;10:1084-96. [PubMed]
- Kelly RS, Vineis P. Biomarkers of susceptibility to chemical carcinogens: the example of non-Hodgkin lymphomas. Br Med Bull 2014;111:89-100. [PubMed]
- Hu Y, Tang H. MicroRNAs regulate the epithelial to mesenchymal transition (EMT) in cancer progression. Microrna 2014;3:108-17. [PubMed]
- Zaccagna F, Anzidei M, Sandolo F, et al. MRgFUS for liver and pancreas cancer treatments: the Umberto I hospital experience. Transl Cancer Res 2014;3:430-41.
- Wang Q, Wei L, Guan X, et al. Briefing in family characteristics of microRNAs and their applications in cancer research. Biochim Biophys Acta 2014;1844:191-7.
- Kaplan BB, Kar AN, Gioio AE, et al. MicroRNAs in the axon and presynaptic nerve terminal. Front Cell Neurosci 2013;7:126. [PubMed]
- Li M, Fu W, Wo L, et al. miR-128 and its target genes in tumorigenesis and metastasis. Exp Cell Res 2013;319:3059-64. [PubMed]
- Leite-Moreira AM, Lourenço AP, Falcão-Pires I, et al. Pivotal role of microRNAs in cardiac physiology and heart failure. Drug Discov Today 2013;18:1243-9. [PubMed]
- Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment Int J Oncol 2013;43:985-94. [PubMed]
- Palumbo S, Miracco C, Pirtoli L, et al. Emerging roles of microRNA in modulating cell-death processes in malignant glioma. J Cell Physiol 2014;229:277-86. [PubMed]
- Ghosh A, Berger A. Opioids, adjuvants, and interventional options for pain management of symptomatic metastases. Ann Palliat Med 2014;3:172-91. [PubMed]
- Francis T, Graf A, Hodges K, et al. Histamine regulation of pancreatitis and pancreatic cancer: a review of recent findings. Hepatobiliary Surg Nutr 2013;2:216-26. [PubMed]
- Wang J, Paris PL, Chen J, et al. Next generation sequencing of pancreatic cyst fluid microRNAs from low grade-benign and high grade-invasive lesions. Cancer Lett 2015;356:404-9. [PubMed]
- Wang J, Raimondo M, Guha S, et al. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. J Cancer 2014;5:696-705. [PubMed]
- Huang X, Lv W, Zhang JH, et al. miR-96 functions as a tumor suppressor gene by targeting NUAK1 in pancreatic cancer. Int J Mol Med 2014;34:1599-605. [PubMed]
- Tu Y, Gao X, Li G, et al. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res 2013;73:6046-55. [PubMed]
- Jin J, Cai L, Liu ZM, et al. miRNA-218 inhibits osteosarcoma cell migration and invasion by down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev 2013;14:3681-4. [PubMed]
- Zhu Z, Xu Y, Du J, et al. Expression of microRNA-218 in human pancreatic ductal adenocarcinoma and its correlation with tumor progression and patient survival. J Surg Oncol 2014;109:89-94. [PubMed]
- Zarogoulidis P, Pavlioglou P, Pivert PL, et al. Current and future intratumoral targeted treatment for pancreatic cancer. Ther Deliv 2014;5:913-26. [PubMed]
- Li X, Truty MA, Kang Y, et al. Extracellular lumican inhibits pancreatic cancer cell growth and is associated with prolonged survival after surgery. Clin Cancer Res 2014;20:6529-40. [PubMed]
- Tholey RM, Lal S, Jimbo M, et al. MUC1 Promoter-Driven DTA as a Targeted Therapeutic Strategy against Pancreatic Cancer. Mol Cancer Res 2015;13:439-48. [PubMed]
- Nakata D. Increased N-glycosylation of Asn88 in serum pancreatic ribonuclease 1 is a novel diagnostic marker for pancreatic cancer. Sci Rep 2014;4:6715. [PubMed]
- Schönhuber N, Seidler B, Schuck K, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med 2014;20:1340-7. [PubMed]
- Sherman MH, Yu RT, Engle DD, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 2014;159:80-93. [PubMed]
- Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev 2012;31:653-62. [PubMed]
- Izzotti A, Cartiglia C, Steele VE, et al. MicroRNAs as targets for dietary and pharmacological inhibitors of mutagenesis and carcinogenesis. Mutat Res 2012;751:287-303. [PubMed]
- Vira D, Basak SK, Veena MS, et al. Cancer stem cells, microRNAs, and therapeutic strategies including natural products. Cancer Metastasis Rev 2012;31:733-51. [PubMed]
- Link A, Kupcinskas J, Wex T, et al. Macro-role of microRNA in gastric cancer. Dig Dis 2012;30:255-67. [PubMed]
- Karsy M, Arslan E, Moy F. Current Progress on Understanding MicroRNAs in Glioblastoma Multiforme. Genes Cancer 2012;3:3-15. [PubMed]
- Shi J, Yang L, Wang T, et al. miR-218 is downregulated and directly targets SH3GL1 in childhood medulloblastoma. Mol Med Rep 2013;8:1111-7. [PubMed]
- Guan H, Wei G, Wu J, et al. Down-regulation of miR-218-2 and its host gene SLIT3 cooperate to promote invasion and progression of thyroid cancer. J Clin Endocrinol Metab 2013;98:E1334-44. [PubMed]
- Palmblad K, Schierbeck H, Sundberg E, et al. High Systemic Levels of the Cytokine-Inducing HMGB1 Isoform Secreted in Severe Macrophage Activation Syndrome. Mol Med 2015;20:538-47. [PubMed]
- Kang R, Chen R, Zhang Q, et al. HMGB1 in health and disease. Mol Aspects Med 2014;40:1-116. [PubMed]
- Srinivasan M, Banerjee S, Palmer A, et al. HMGB1 in hormone-related cancer: a potential therapeutic target. Horm Cancer 2014;5:127-39. [PubMed]
- Zhou TB. Role of high mobility group box 1 and its signaling pathways in renal diseases. J Recept Signal Transduct Res 2014;34:348-50. [PubMed]
- Chen R, Hou W, Zhang Q, et al. Emerging role of high-mobility group box 1 (HMGB1) in liver diseases. Mol Med 2013;19:357-66. [PubMed]
- Musumeci D, Roviello GN, Montesarchio D. An overview on HMGB1 inhibitors as potential therapeutic agents in HMGB1-related pathologies. Pharmacol Ther 2014;141:347-57. [PubMed]
- Abe A, Kuwata T, Yamauchi C, et al. High Mobility Group Box1 (HMGB1) released from cancer cells induces the expression of pro-inflammatory cytokines in peritoneal fibroblasts. Pathol Int 2014;64:267-75. [PubMed]
- Ueda M, Takahashi Y, Shinden Y, et al. Prognostic significance of high mobility group box 1 (HMGB1) expression in patients with colorectal cancer. Anticancer Res 2014;34:5357-62. [PubMed]
- Parker KH, Sinha P, Horn LA, et al. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res 2014;74:5723-33. [PubMed]
- Zuo Z, Che X, Wang Y, et al. High mobility group Box-1 inhibits cancer cell motility and metastasis by suppressing activation of transcription factor CREB and nWASP expression. Oncotarget 2014;5:7458-70. [PubMed]
- Meng Q, Zhao J, Liu H, et al. HMGB1 promotes cellular proliferation and invasion, suppresses cellular apoptosis in osteosarcoma. Tumour Biol 2014;35:12265-74. [PubMed]
- Wang W, Zhu H, Zhang H, et al. Targeting HMGB1 inhibits bladder cancer cells bioactivity by lentivirus-mediated RNA interference. Neoplasma 2014;61:638-46. [PubMed]


