A PTEN translational isoform has PTEN-like activity
Introduction
PTEN is one of the most frequently lost or mutated tumor suppressors in human cancers (1-6). Germline mutations of PTEN are often associated with cancer predisposition syndromes, such as Cowden disease, which is characterized by multiple hamartomas (7). The role of PTEN as a potent tumor suppressor has been validated in various mouse gene targeting and transgenic models, where deletion or inactivation of PTEN led to development of different types of tumors that mimic human cancers (8-13), whereas gain of extra PTEN induced a tumor-suppressive metabolic state (14). PTEN loss also results in neurological disorders, metabolic diseases, and tissue homeostasis defects (15-20), suggesting that PTEN has functions other than tumor suppression. PTEN is also essential for embryonic development as homozygous deletion of PTEN in mice resulted in developmental failure and eventually embryonic lethality (8). These findings demonstrate the PTEN is important in diverse biological processes including tumor suppression, development, tissue homeostasis, and metabolism (21).
At cellular and biochemical levels, PTEN plays critical roles in cell proliferation, survival, death and migration (22-27), principally by antagonizing PI3K-Akt dependent signaling through its lipid phosphatase activity (23,28-31). PTEN dephosphorylates PIP3 at the cell membrane and negatively regulates PI3K-Akt mediated signaling. PTEN also has protein phosphatase and phosphatase-independent activities that are not related to the PI3K-Akt pathway (24,32-34), indicating that the roles of PTEN plays in diverse fundamental biological processes cannot be attributed only to its lipid phosphatase activity. To date, some observed consequences resulted from loss of PTEN are pending to explain. Like many other tumor suppressors and oncoproteins, unidentified isoforms of PTEN may exist to serve in roles previously attributed to canonical PTEN.
In this study, we identified a new translation initiation at a CUG site in the 5' untranslated region (5' UTR) of PTEN mRNA. Translation from this CUG start codon generates a larger isoform of PTEN with an extended N-terminal region of an extra 173 amino acids. We showed this PTEN isoform, similar to PTEN, has lipid phosphatase activity. In addition, overexpression of PTEN isoform inhibited cell growth, induced Caspase-mediated apoptosis, and inhibited cell migration, indicating this isoform of PTEN is another tumor suppressor.
Material and methods
Western blotting
Cell lysates were prepared in radioimmunoprecipitation assay (RIPA) buffer (50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 0.5% deoxycholate, 0.1% SDS and 1.0% NP-40) (Thermo Scientific, Rockford, IL, USA) containing a protease inhibitor cocktail. Total protein of 50 µg per well was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes (PVDF, Immobilon P, Millipore, Bedford, MA, USA) for Western blotting. Western blotting was carried out using standard protocols. In brief, the membrane was blocked in 5% low fat milk powder in PBST (3.2 mmol/L Na2HPO4, 0.5 mmol/L KH2PO4, 1.3 mmol/L KCl, 135 mmol/L NaCl, 0.05% Tween-20, pH 7.4) for 30 min. The primary antibodies against indicated proteins were incubated either overnight at 4 °C or 2 h at room temperature. After wash, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Santa Cruz Biotechnology) for 20 min at room temperature. The bound antibodies were visualized by an enhanced chemiluminescence (ECL) detection system. Antibodies against PTEN were purchased from CASCADE BioScience (Winchester, MA, USA) (C-terminal epitope) and ABGENT (WuXi, China) (N-terminal epitope). Akt, pAkt (Ser473), Cleaved Caspase 3, S6K, and pS6K (Thr389) were from Cell Signaling (Danvers, MA, USA), whereas Actin was from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell culture and transfection
NIH3T3, LNCaP, HeLa, COS-1, PC3, MCF-7, U2OS, MDA1386, H1299, DU145, and 293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM high glucose medium supplemented with 10% of fetal bovine serum (FBS), 50 mg/mL penicillin and 50 mg/mL streptomycin (Invitrogen, Carlsbad, CA, USA). Cells were maintained in an incubator with a humidified atmosphere of 95% and 5% CO2 at 37 °C. For transfection, cells were transiently transfected with indicated constructs using Lipofectamin 2,000 transfection reagent (Life Technology, Carlsbad, CA, USA) according to the manufacturer’s protocol. In brief, cells were seeded in 100 mm dishes (Nunc, Roskilde, Denmark) 24 h before transfection and transiently transfected at a confluency of 80-90%. The transfection mixture was diluted in Opti-MEM serum-free media (Life Technology, Carlsbad, CA, USA) and cells were incubated in Opti-MEM serum-free media as well. Six hours after transfection, the transfection media was changed to the complete media containing 10% FBS and antibiotics. Cells were either harvested 48 h after transfection for protein extraction or selected with Geneticin 24 h after transfection to establish stable cells. The shRNA sequences against human PTEN were 5'-CCACAAATGAAGGGATATAAACTCGAGTTTATATCCCTTCATTTGTGG-3' (3UTR), 5'-ATTTCGGGCACCGCATATTAACTCGAGTTAATATGCGGTGCCCGAAA-3' (3UTR), and 5'-CTAGAACTTATCAAACCCTTTCTCGAGAAAGGGTTTGATAAGTTCTAG-3' (CDS). 5'-CGTGCAGATAATGACAAGGAACTCGAGTTCCTTGTCATTATCTGCACG-3' (CDS), and 5'-AGGCGCTATGTGTATTATTATCTCGAGATAATAATACACATAGCGCC-3' (CDS).
Immunoprecipitation
Cells were lysed as described above. Lysate was incubated with 3 µg of PTEN antibody plus 20 µL of protein an agarose beads (GE Healthcare Life Sciences, Pittsburg, PA, USA) by gentle rocking at 4 °C overnight. The beads were then washed three times with lysis buffer and one time with PBS. Bound proteins were resuspended in 2X Laemmli buffer, separated by SDS-PAGE gels, and analyzed by Western blotting.
Proliferation and apoptosis assay
For proliferation assays, 0.2×106 of cells stably expressing indicated proteins were seeded, cells were harvested daily, and cell numbers were counted, in triplicates. For apoptosis assay, cells stably expressing indicated proteins were grown to 60% confluence in complete media before switching to serum free media. Cells were then harvested after 48 h and subjected to Caspase 3 activity assay using a kit from Cell Signaling Technology. Basically, the kit contains a fluorogenic substrate (N-Acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin or Ac-DEVD-AMC) for Caspase 3. During the assay, activated Caspase 3 cleaves this substrate between DEVD and AMC, generating highly fluorescent AMC that can be detected using a fluorescence reader with excitation at 380 and emission at 445 nm. Cleavage of the substrate occurs in lysates of apoptotic cells, therefore the amount of AMC produced is proportional to the number of apoptotic cells in the sample. For UV irradiation, cells stably expressing indicated proteins were irradiated at 300 milliJoules using GS Gene Linker (Bio-Rad) and were followed by imaging analysis.
Migration assays
Cell migration was performed as previously described (35) with slight modifications. In brief, cells stably expressing indicated proteins were grown to subconfluent on culture dishes. The cells were starved of serum for 24 h and treated with Mitomycin C for 90 min to arrest cell proliferation. A wound track was then introduced by scraping the cell monolayer with a thin pipette tip. Cells migrating into the wound area were then followed with phase-contrast microscope for up to 16 h.
Results
PTEN isoform is not PTEN post-translational modification
When we assessed PTEN protein levels in different cell lines in which PTEN is either intact, mutated, or deleted, we found the antibodies for PTEN recognized a protein, about 15 kDa bigger than canonical PTEN, that co-existed with PTEN. This protein was recognized by antibodies raised against epitopes of both N-terminal and C-terminal amino acids of PTEN and its abundance is proportional to that of PTEN (Figure 1A), indicating it might be a PTEN post-translational modification or PTEN isoform from RNA alternative splicing.
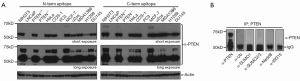
Based on the difference in size between PTEN and its isoform, we speculated that PTEN isoform is a type of post-translational modification involving addition of small protein or peptides. To assess such possibility, we immunoprecipitated PTEN and its isoform from 293T cell lysate by antibody against PTEN and subject immunoprecipitated to Western blotting analysis with antibodies against PTEN, ISG15, SUMO1/2/3, Nedd8, or ubiquitin. Western blotting using PTEN antibody showed both PTEN and its isoform were successfully enriched by immunoprecipitation. However, PTEN isoform was not recognized by any other antibodies, indicating it’s not a common product of ubiquitin or ubiquitin-like modification (Figure 1B).
PTEN isoform is a translational variant of PTEN
To further evaluate the possibility whether PTEN isoform is a type of posttranslational modification of PTEN, we transfected PTEN cDNA with either N-terminal or C-terminal HA tag into 293T cells and analyzed expression of PTEN and its isoform by Western blotting. We found, compared with PTEN that was expressed at high levels, PTEN isoform was not detectable at all, suggesting PTEN isoform is not from posttranslational modification of PTEN (Figure 2A).
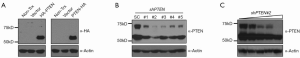
To test whether PTEN isoform is a translational variant of PTEN or a product from RNA alternative splicing, we knocked-down PTEN in 293T cells with sequences targeting coding sequences or 3'-UTR of PTEN. Cells with PTEN knocked-down were lysed and analyzed with Western blotting with antibody against PTEN. As shown in Figure 2B, the protein levels of PTEN in 293T cells were reduced to different extents by different targeting sequences, compared with scramble control sequence transfected cells. The expression of PTEN isoform was also reduced, to the extents proportional to the levels of PTEN. Furthermore, transfection of gradient amounts of shRNA of PTEN into 293T cells resulted in gradient reduction in both PTEN and PTEN isoform (Figure 2C). These results indicated PTEN isoform is either a translational variant of PTEN or a product resulting from RNA alternative splicing and the sequences of its mature mRNA include coding sequence and 3'-UTR of PTEN. Bioinformatic analysis of PTEN mRNA disclosed an alternative starting code (CUG513), 5' upstream of the coding sequence of PTEN, which generates a predicted protein at the same size as observed PTEN isoform.
PTEN isoform has PTEN-like activity to inhibit PI3K-Akt pathway
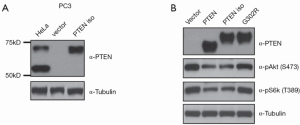
Based on the identified alternative starting code, we cloned human PTEN isoform. To study PTEN isoform-specific function and avoid co-expression of PTEN, we replaced its starting code CUG with AUG and mutated PTEN’s canonical starting code from AUG to GCG (Ala) in PTEN isoform cDNA. Transfection of the cDNA of PTEN isoform into PC3 (a PTEN-null cell line) cells resulted in expression of a protein that migrated to the position of endogenous PTEN isoform from HeLa (Figure 3A). To begin to assess biochemical activity of PTEN isoform, we subjected cell lysates of PC3 cells transfected with PTEN isoform to Western blot analysis using antibodies against PTEN, pAkt (Ser473), pS6K (Thr389), and total Akt. As shown in Figure 3B, the levels of phosphorylated Akt and S6K were reduced by more than 50% in cells expressing PTEN or PTEN isoform, compared to cells transfected with empty vector, indicating PTEN isoform, similar to PTEN, is able to negatively regulate PI3K-Akt signaling. To prove it, we created a missense mutation, G302 to R, in the phosphatase domain of PTEN isoform, which is analogous to G129 to R in PTEN—a mutation that is depleted of lipid phosphatase activity (30). We found G302R mutant failed to inhibit Akt phosphorylation (Figure 3B, lanes 3). Together, the results suggested PTEN isoform is able to suppress PI3K-Akt signaling pathway in a lipid phosphatase-dependent manner.
Overexpression of PTEN-Long inhibits cell proliferation and induces cell death
PTEN is well known to play a crucial role in cell proliferation and death by regulating the levels of PIP3 that activates Akt, a central regulator of survival and death of cell. To understand the role that PTEN isoform possibly plays in regulating cell proliferation and cell death, we stably overexpressed PTEN, PTEN isoform, or PTEN isoform (G302R) in HepG2 cells and assessed their effects on cell growth and induction of cell death. We found overexpression of PTEN or PTEN isoform in HepG2 cells inhibited cell proliferation, as compared to cells transfected with empty vector alone (Figure 4A,B). In addition, this activity depends on PTEN isoform's lipid phosphatase activity as PTEN isoform (G302R) showed no effect (Figure 4B).
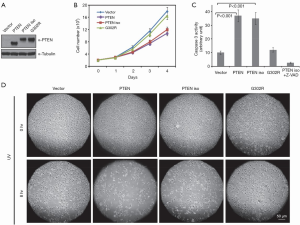
To understand the role of PTEN isoform in cell death, HepG2 cells stably expressing PTEN isoform or PTEN isoform (G302R) were serum-starved for 48 h and then subjected to Caspase 3 activity assay. As shown in Figure 4C, a total of 48 h of serum starvation increased Caspase 3 activity in PTEN or PTEN isoform-expressing HepG2 cells by more than 2-fold, as compared to empty vector or PTEN isoform (G302R) expressing cells (Figure 4C), indicating PTEN isoform has apoptotic activity and this apoptotic activity is dependent on its lipid phosphatase activity. We also subjected HepG2 cells stably expressing indicated proteins to UV irradiation and assessed their effects on UV irradiation induced apoptosis. The results showed overexpression of PTEN isoform also sensitizes cells to UV irradiation-induced apoptosis and its lipid phosphatase activity played an essential role (Figure 4D).
Overexpression of PTEN-Long inhibits cell migration
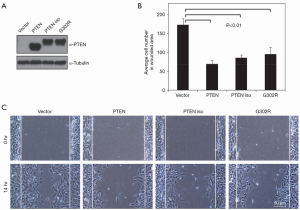
Next, we investigated the contribution of PTEN isoform to the migratory properties of PTEN knockout MEF cells by scratch assay. Cells stably expressing PTEN, PTEN isoform, or its mutant were allowed to grow confluent before being scratched and monitored with microscope. Cell migration was assessed by the speed of wound closure. As shown in Figure 5A,B, expression of PTEN isoform inhibited migration of cells, as compared cells transfected with empty vector alone (Figure 5B). Quantitative analysis of cells migrated into wound area 14 h after scratching revealed a more than 50% (P=0.01) inhibition in migration of PTEN knockout MEF cells into the wound track by expression of PTEN or PTEN isoform (Figure 5C). Interestingly, overexpression of the lipid phosphatase-dead mutant of PTEN isoform also inhibited cell migration, to the extent similar to wild type PTEN isoform. Because G302 to R mutant retains protein phosphatase activity, inhibition of cell migration by the G302R mutant indicates that lipid phosphatase activity is not essential for PTEN isoform’s impact on cell migration.
Discussion
Although the tumor suppressor PTEN has been intensively studied since its discovery back in 1997, some fundamental aspects of its functions are still unclear. Seventeen years ago PTEN was claimed to possess protein phosphatase activity, but the first proved substrate was reported only recently (32). Evidence indicated some roles previously assumed to belong to PTEN might be attributed to unidentified PTEN isoform. In eukaryotes, protein translation is most commonly started at AUG codons, and the efficiency of initiation also depends on the nucleotides before and after start codon. Recently, it was reported that initiation of protein translation also occurs at non-AUG codons (36-39). These alternate initiation codons were believed to increase both genome coding capacity and protein diversity. The identification of PTEN isoform advances our understanding of diversity of PTEN family proteins mediated by alternative translation initiation.
Recently, Hopkins and colleagues identified a translational variant of PTEN (PTEN-Long) that is secreted and can enter other cells (40). Like PTEN, PTEN-Long antagonized the PI3K-Akt pathway and induced cell death. Treatment with PTEN-Long led to tumor regression in several mouse models, indicating it might have therapeutic potential. Soon after, Liang and colleagues reported the identification of PTENα, a PTEN isoform that is localized to mitochondria, where it collaborates with PTEN to regulate mitochondrial energy metabolism (41). PTENα is also translated by an alternative, CUG-dependent mechanism that is similar to PETN-Long. Because PTEN-Long and PTENα are involved in different biological processes, they are possibly two different PTEN isoforms and CUG-dependent translation contributes to protein diversity in eukaryotes.
As PTEN isoform includes the whole PTEN sequences, phenotype seen from previously created Pten knockout mice were essentially consequence of Pten and Pten isoform double knockout. Although the PTEN proteins are more abundant than PTEN isoform, phenotypic deficiencies in these double knockout mice are at least partially attributable to the loss of Pten isoform. In order to dissect functions of PTEN from PTEN isoform, it is possible to create Pten isoform-specific knockout mouse by use of its unique nucleotide sequences N-terminal to PTEN and assess loss-of-function by comparing single and double knockout models. These models will also help to re-evaluate contribution of each PTEN family proteins to tumorigenesis, embryonic development, and other biological processes. Meanwhile, as PTEN isoform has extra amino acids N-terminal to PTEN sequence, bioinformatics analysis of these extended sequences might predict function and regulation of PTEN isoform. This region could contain signaling sequences and/or conserved domain which endow PTEN isoform distinct cellular localization and functions that do not overlap PTEN lipid or protein phosphatase activities. It should be noted that expression of PTEN isoform is generally much lower than PTEN. It is therefore interesting to investigate whether functions of PTEN and PTEN isoform are complementary or synergistic. Furthermore, PTEN is a common target of mutations in various types of cancers. These mutations span PTEN’s coding and noncoding regions. It is also interesting to know whether PTEN isoform has specific N-terminal mutation(s) outside of PTEN sequences that are cancer- and/or other diseases-relevant. Further studies are needed to show whether and how PTEN isoform participates in distinct cellular processes and whether PTEN family has other members so far unidentified.
Acknowledgement
Funding: Advanced key Scientific and Technological Programs of Ningbo (2013C51009) and National Natural Science Foundation of China (31271451).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997;275:1943-7. [PubMed]
- Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 1997;15:356-62. [PubMed]
- Teng DH, Hu R, Lin H, et al. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res 1997;57:5221-5. [PubMed]
- Wang SI, Puc J, Li J, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res 1997;57:4183-6. [PubMed]
- Whang YE, Wu X, Suzuki H, et al. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci U S A 1998;95:5246-50. [PubMed]
- Liu C, Li G, Chen R, et al. A novel PTEN gene promoter mutation and untypical Cowden syndrome. Chin J Cancer Res 2013;25:306-11. [PubMed]
- Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 1997;16:64-7. [PubMed]
- Di Cristofano A, Pesce B, Cordon-Cardo C, et al. Pten is essential for embryonic development and tumour suppression. Nat Genet 1998;19:348-55. [PubMed]
- Podsypanina K, Ellenson LH, Nemes A, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A 1999;96:1563-8. [PubMed]
- Kimura T, Suzuki A, Fujita Y, et al. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development 2003;130:1691-700. [PubMed]
- Suzuki A, Itami S, Ohishi M, et al. Keratinocyte-specific Pten deficiency results in epidermal hyperplasia, accelerated hair follicle morphogenesis and tumor formation. Cancer Res 2003;63:674-81. [PubMed]
- Tsuruta H, Kishimoto H, Sasaki T, et al. Hyperplasia and carcinomas in Pten-deficient mice and reduced PTEN protein in human bladder cancer patients. Cancer Res 2006;66:8389-96. [PubMed]
- Yoo LI, Liu DW, Le Vu S, et al. Pten deficiency activates distinct downstream signaling pathways in a tissue-specific manner. Cancer Res 2006;66:1929-39. [PubMed]
- Garcia-Cao I, Song MS, Hobbs RM, et al. Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell 2012;149:49-62. [PubMed]
- Gasser T. Update on the genetics of Parkinson's disease. Mov Disord 2007;22 Suppl 17:S343-50. [PubMed]
- Stiles B, Wang Y, Stahl A, et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity Proc Natl Acad Sci U S A 2004;101:2082-7. [corrected]. [PubMed]
- Stiles BL, Kuralwalla-Martinez C, Guo W, et al. Selective deletion of Pten in pancreatic beta cells leads to increased islet mass and resistance to STZ-induced diabetes. Mol Cell Biol 2006;26:2772-81. [PubMed]
- Nguyen KT, Tajmir P, Lin CH, et al. Essential role of Pten in body size determination and pancreatic beta-cell homeostasis in vivo. Mol Cell Biol 2006;26:4511-8. [PubMed]
- Suzuki A, Kaisho T, Ohishi M, et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med 2003;197:657-67. [PubMed]
- Kishimoto H, Ohteki T, Yajima N, et al. The Pten/PI3K pathway governs the homeostasis of Valpha14iNKT cells. Blood 2007;109:3316-24. [PubMed]
- Kishimoto H, Hamada K, Saunders M, et al. Physiological functions of Pten in mouse tissues. Cell Struct Funct 2003;28:11-21. [PubMed]
- Li J, Simpson L, Takahashi M, et al. The PTEN/MMAC1 tumor suppressor induces cell death that is rescued by the AKT/protein kinase B oncogene. Cancer Res 1998;58:5667-72. [PubMed]
- Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998;95:29-39. [PubMed]
- Tamura M, Gu J, Matsumoto K, et al. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 1998;280:1614-7. [PubMed]
- Yamada KM, Araki M. Tumor suppressor PTEN: modulator of cell signaling, growth, migration and apoptosis. J Cell Sci 2001;114:2375-82. [PubMed]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell 2000;100:387-90. [PubMed]
- Su X, Xing J, Wang Z, et al. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res 2013;25:235-9. [PubMed]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 1998;273:13375-8. [PubMed]
- Wu X, Senechal K, Neshat MS, et al. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A 1998;95:15587-91. [PubMed]
- Myers MP, Pass I, Batty IH, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A 1998;95:13513-8. [PubMed]
- Sun H, Lesche R, Li DM, et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci U S A 1999;96:6199-204. [PubMed]
- Shi Y, Wang J, Chandarlapaty S, et al. PTEN is a protein tyrosine phosphatase for IRS1. Nat Struct Mol Biol 2014;21:522-7. [PubMed]
- Shen WH, Balajee AS, Wang J, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007;128:157-70. [PubMed]
- Song MS, Carracedo A, Salmena L, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 2011;144:187-99. [PubMed]
- Sano S, Itami S, Takeda K, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J 1999;18:4657-68. [PubMed]
- Hann SR, King MW, Bentley DL, et al. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell 1988;52:185-95. [PubMed]
- Malarkannan S, Horng T, Shih PP, et al. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity 1999;10:681-90. [PubMed]
- Németh AL, Medveczky P, Tóth J, et al. Unconventional translation initiation of human trypsinogen 4 at a CUG codon with an N-terminal leucine. A possible means to regulate gene expression. FEBS J 2007;274:1610-20. [PubMed]
- Gerashchenko MV, Su D, Gladyshev VN. CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. J Biol Chem 2010;285:4595-602. [PubMed]
- Hopkins BD, Fine B, Steinbach N, et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 2013;341:399-402. [PubMed]
- Liang H, He S, Yang J, et al. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 2014;19:836-48. [PubMed]


