Overexpression of Gli1 in cancer interstitial tissues predicts early relapse after radical operation of breast cancer
Objective: To investigate whether Gli1 expression is important in relapse after radical operation of breast
cancer.
Methods: Using immunohistochemistry, Gli1 expression was analyzed in human primary breast cancer
(n=284) and paracancerous tissues (n=20), and also in local lymph nodes (n=28) and metastatic lymph nodes
(n=28).
Results: Initial analysis of Gli1 expression in a small cohort of 20 breast tumors and their paracancerous
tissues showed a tendency towards Gli1 overexpression in breast cancer tissues (P<0.001). Further, Gli1
expression in 284 breast cancer tissue samples was analyzed and a significant correlation was found between
increased expression of nuclear Gli1 and unfavorable recurrence-free survival (RFS) (P<0.05). The nuclear
expression of Gli1 in metastatic lymph nodes following relapse after radical operation was much higher than
that in the local lymph nodes of primary carcinoma (P<0.05). Most interestingly, the expression of Gli1 was
much higher in the interstitial tissues of the relapsed group than of the non-relapsed group (P<0.001).
Conclusions: Breast cancer shows a high prevalence of Gli1 expression, which is significantly correlated
with aggressive features and unfavorable RFS. Nuclear Gli1 overexpression, especially in the interstitial
tissues, signified early relapse after radical operation of breast cancer.
Key words: Gli1; prognosis; breast cancer
Introduction
Breast cancer is a significant global public health problem. Despite major advances in adjuvant therapy, therapeutic options for patients with unresectable metastatic disease, recurrent disease or triple negative breast cancer are limited. Approximately 39,840 patients died of this disease in the USA in 2010 (1), emphasizing the need for new therapies. A better understanding of the mechanisms that underlie the development of breast cancer would help to identify novel molecular targets for its treatment and determine more accurate clinicopathological prognostic factors.
Hedgehog (HH) is a major pathway that is critical in embryonic development (2,3), stem cell maintenance (4) and tumorigenesis (5). It not only contributes to development and regeneration, but also leads to transformation of progenitors when continuously activated, which might culminate in cancers such as small-cell lung carcinoma (6) and prostate (7) and digestive tract tumors (8). Most interestingly, aberrant reactivation of HH signaling in breast cancer has been reported (9,10). Gli proteins act at the downstream end of the HH pathway where they control transcriptional programs in response to pathway activation. Indeed, the induction of Gli1 mRNA expression by HH signaling is a reliable marker for pathway activity (11).
There are no reported data on the dynamic changes of Gli1 expression, which is a key HH pathway target, in breast cancer patients who experienced relapse/metastasis. The current study retrospectively investigated the expression of the Gli1 protein in human breast cancer tissues and evaluated the clinical implications of HH signal activation for patients who experienced relapse or metastasis.
Patients and methods
Patients and tissue samples
A total of 302 breast cancer patients (median age, 52 years; range, 26-78 years) with positive pathological examination findings (including ductal and lobular cancer) were all chosen from the Second Affiliated Hospital of Dalian Medical University, Dalian, China between April 2002 and December 2004. Of the 302 breast cancer samples, 18 cases with missing records of survival time and survival status were excluded. The clinicopathological characteristics and the outcomes of these patients were recorded. Forty-five patients were suffering from cancer relapse, of whom 28 cases had metastatic superficial nodules confirmed by pathology, and the metastatic superficial nodules were named “metastatic lymph nodes”. In comparison, the lymph nodes identified during the radical operation were named “local lymph nodes”, and these 45 patients formed the ‘relapsed group’. The other 239 cases had not experienced relapse by the end of the follow-up period and they formed the ‘non-relapsed group’. The observation period was from 2002 to 2009, with a median follow-up time of 62 months (range, 3-83 months). Clinical and pathologic factors were evaluated, including age, T- and N-classification, clinical stage, presence of steroid receptors and human epidermal growth factor receptor 2 (HER-2) expression. To use these clinical materials for research purposes, patients’ written consent and the approval from the Ethic Committee of the Second Affiliated Hospital of Dalian Medical University were obtained before the study. The clinical stages of all the patients were classified according to the 2002 TNM staging of the International Union Against Cancer (UICC). Follow-up information was obtained from office charts, hospital records, and telephone interviews.
Immunohistochemistry
Immunohistochemical analysis was carried out using the HistostainTM-Plus Kit (ZSGB-Bio, Beijing, China) according to the manufacturer’s instruction. Paraffin-embedded breast cancer sections (4 μm) were deparaffinized with xylene and rehydrated. Subsequently, antigen retrieval was performed by heating in an autoclave in 10 mmol/L sodium citrate buffer (pH 6.0). Then the sections were treated with 3% hydrogen peroxide in methanol to quench the endogenous peroxidase activity, and 1% bovine serum albumin was used to block non-specific binding, followed by incubation of the sections with a rabbit polyclonal anti-Gli1 antibody (1:100, Santa Cruz sc-20687, CA) overnight at 4 ℃. After washing with phosphate buffered saline (PBS), the sections were incubated with biotinylated secondary antibody, followed by incubation with the streptavidin-horseradish peroxidase complex. The sections were then immersed in 3,3'-diaminobenzidine (DAB) for 6 min, counterstained with 10% Mayer’s hematoxylin, dehydrated, and mounted in crystal mount. PBS was used instead of primary antibody for negative control. To minimize variation in the immunopositive cells, all sections were stained in DAB for the same amount of time. Two pathologists, blinded to the clinical outcome, scored the results of the staining independently. Measurements of estrogen receptor (ER), progesterone receptor (PR) and HER-2 were routinely performed as previously described (12).
Evaluation of immunohistochemistry
Gli1 expression both in interstitial tissues of cancer and in cancer tissues was assessed as either high or low by using the Immuno-Reactive-Score (IRS). First, the number of positive cells and staining intensity were evaluated semi-quantitatively. The IRS is the product of the staining intensity (graded between negative =0 and strong =3) and the percentage of positively stained cells (graded between 0 and 4; 1≤25%, 2=25-50%, 3=51-75%, and 4≥75%). If the final IRS score was 7-12, Gli1 expression was considered high, otherwise Gli1 expression was considered low (13). Cases with discrepancies in IRS score were discussed together with other pathologists until a consensus was reached.
For evaluation of ER and PR, the fraction of positively staining nuclei was categorized into negative (0-1%), weak (2-10%), medium (11-75%) or high (76-100%) (12). For statistical analyses, variables were assigned as “negative” or “positive” using the clinically established cut-off for ER and PR at 10% positive nuclei. The evaluation of HER-2 was performed according to current clinical practice (14). Amplification of HER-2 gene in tumors with a medium HER-2 staining intensity of “score 2” was tested by additional fluorescence in situ hybridization (FISH) analyses in order to determine HER-2 amplification of these tumors.
Statistical analysis
Immunohistochemical data were statistically analyzed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). P<0.05 was defined to be statistically significant. The Chi-Square test was employed to evaluate the differences in Gli1 expression between the different categories of tissues and to assess the correlation between clinical features and Gli1 expression in breast cancer. The score of immunoreactivity was analyzed and compared by independent t-tests. Recurrence-free survival (RFS) was calculated in accordance with the Kaplan-Meier equation and compared using the log-rank test.
Results
Increased expression of Gli1 in primary breast cancer tissues
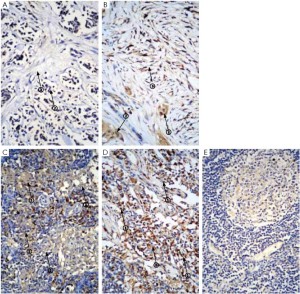
We analyzed Gli1 expression in a set of tumor and adjacent tissues (n=20 respectively, Table 1). Gli1 could be detected in both the nuclei and cytoplasm of non-cancerous cells, but was mainly localized to the nuclei of cancer cells. Because Gli1 is a transcription factor, nuclear Gli1 expression was scored in subsequent analyses. Among the 20 samples, 17 exhibited high levels of Gli1 expression in the tumor tissues, whereas 5 of the 20 paracancerous tissues displayed high expression levels. The tumors (Figure 1A, B) exhibited a higher level of Gli1 expression, and the overexpression of Gli1 was statistically significant (P<0.001).

Full table
Correlation analysis of Gli1 expression and clinicopathological parameters
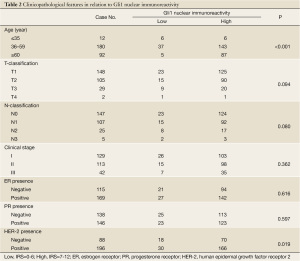
We further analyzed the Gli1 expression of 284 breast cancer tissue samples and examined possible correlations between Gli1 expression profiles and the patients’ clinicopathological characteristics. As presented in Table 2, Gli1 expression was strongly correlated with age (P<0.001) and HER-2 expression (P=0.019). No significant association was found between Gli1 expression and T-classification (P=0.094), N-classification (P=0.08), clinical stage (P=0.362), or the presence of ER (P=0.616) or PR (P=0.597).
Full table
Gli1 nuclear overexpression predicts relapse
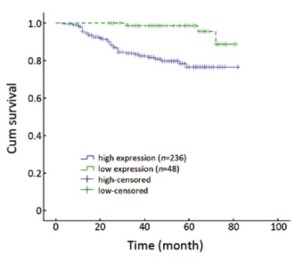
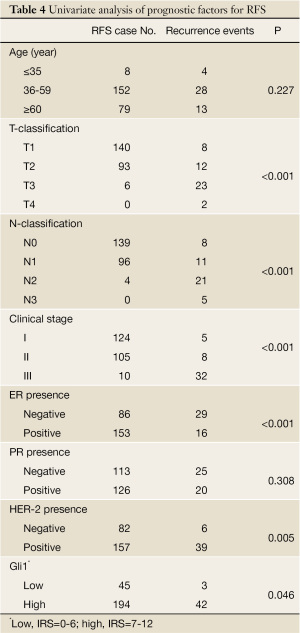
We analyzed the relationship between Gli1 nuclear overexpression and RFS. As expected, Gli1 expression displayed a significant correlation with RFS (P=0.046, Table 3). The recurrence rate, assessed by the Kaplan-Meier method (Figure 2), was 17.8% (42/236) in the high expression group, whereas it was only 6.2% (3/48) in the low expression group (P=0.046). The clinicopathological classification (T- and N- classification), ER and HER-2 status, and clinical stage were important indicators for RFS in breast cancer (P<0.05, respectively), but age and the presence of PR were not (Table 4).

Full table
Full table
To further assess the factors affecting RFS, we stratified patients into two various clinicopathological categories, and separated them based on Gli1 expression (shown in Table 5). The odds ratio (OR) was determined and the 95% confidence interval (95% CI) was estimated. These are shown in Table 6. After stratification, T- and N-classification, clinical stage and ER seemed to serve as a risk factor (P<0.001, P<0.001, P<0.001, and P=0.002, respectively) of breast cancer relapse in the Gli1 high expression group, age could in the low expression group (P=0.016), clinical stage could in either group, while PR could in neither group (P>0.05, respectively).
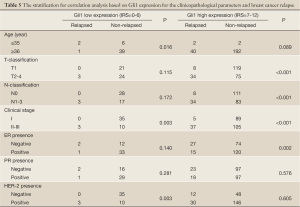
Full table
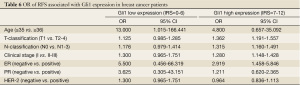
Full table
The recurrence rate was 17.8% (42/236) in the high expression subset compared to 6.3% (3/48) in the low expression subset. However, when only patients with T2+3+4 classifications were analyzed, the recurrence rate was 31.2% (34/109) in the high expression group and 11.1% (3/27) in the low expression group. A similar result was found in patients with N1+2+3 and clinical stages II+III, which were also shown in Kaplan-Meier curves of RFS (Figure 3). That is, the recurrence rate was much higher in the high expression group than in the low expression group. Furthermore, in our study, the median RFS was 25 months (range, 3-67 months) in the Gli1 high expression group compared to 64 months (range, 8-83 months) in the Gli1 low expression group.

Gli1 overexpression in metastatic lymph nodes of relapsed breast cancers
Gli1 was mainly detected in the nuclei of leukomonocytes in the lymph nodes (Figure 1C,D) of breast cancer patients which was much higher than non-metastatic lymph nodes (Figure 1E), and 21/28 local lymph nodes examined exhibited increased levels of Gli1 expression compared to 27/28 metastatic lymph nodes. The scores for the proportion of positively staining cells and the staining intensity of Gli1 in the nuclei of the samples from local lymph nodes of primary carcinoma were not significantly different from the scores for samples taken from metastatic lymph nodes following relapse after radical operation (P=0.099, P=0.16, respectively, Figure 4). However both the proportion of samples with a high IRS score and the absolute value of the IRS score in the metastatic lymph nodes were higher than those in the local lymph nodes, which was statistically significant (P=0.022, P=0.039, respectively, Table 7, Figure 4).
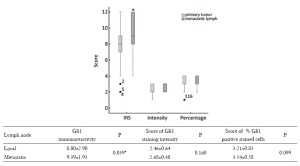

Full table
Gli1 overexpression in interstitial tissues is associated with relapse after radical operation of breast cancer
We found Gli1 could be detected either in interstitial tissues of cancer or in cancer tissues, or in both. In the interstitial tissues of primary cancer, the rate of Gli1 overexpression was 89.3% (25/28). The rate was 96.4% (27/28) in the cancer tissue samples from the relapsed group. In contrast, it was 13.4% (32/239, P<0.001) in the interstitial tissues and 81.2% (194/239) in the cancer tissues in the non-relapsed group. The difference was statistically significant (P<0.05, Table 8). The median RFS was 16 months (range, 3-58 months) in the Gli1 high expression group in contrast to 66 months (range, 8-76 months) in the Gli1 low expression group.

Full table
Discussion
Although lymph node metastasis, large tumor size, and poorly differentiated histopathological grade are commonly considered to be established prognostic markers of metastasis (15), distant metastasis still occurs and the processes of invasion and metastasis that cause mortality in patients are extraordinarily distinctive features of breast cancer progression (16). Thus, it is imperative to address the factors underlying tumorigenesis, invasion and metastasis.
The name of Hedgehog is derived from the appearance of the mutant fruit fly embryo, which is covered with bristles. The HH genes are highly conserved and exist in many other organisms and mammals. Gli is a member of HH signaling cascade and was first described in 1980 through the seminal work of Nüsslein-Volhard and Wieschaus (17). Vertebrate HH genes were subsequently reported in 1993 (18-21). In common with other pathways, HH is mainly composed of three parts: ligands (HH: DHH, IHH, and SHH), receptor complexes and transcription factors. HH proteins act as ligands for receptor complexes located on the membrane of nearby cells. The mammalian HH receptor complex consists of at least three transmembrane proteins: patched 1 protein (PTCH1), Smoothened (SMO) and HH interacting protein (HIP) (22,23). In the absence of ligands, PTCH1 binds to SMO and blocks its function. In the presence of ligands, SMO initiates a signaling cascade that results in activation of the transcription factors Gli1, Gli2, and Gli3 (24). Gli proteins that serve as nuclear executors of HH-signaling contain a C-terminal transactivation domain, but Gli2 and Gli3 were shown to have an N-terminal repression domain. Modification of the N-terminus of Gli2 and Gli3 may be a critical step in the regulation of the transcriptional activity of Gli proteins. Removal of a large portion of the N-terminus results in constitutively active Gli2 and Gli3 proteins (25). In contrast to Gli2 and Gli3, Gli1 is mainly regulated at the transcriptional level and represents a direct transcriptional target of the HH pathway (26-29). Gli1 expression, especially after nuclear translocation, is considered a valuable marker of HH activation (30-35). Activated HH allows the translocation of Glis (Gli1, 2, 3) to the nucleus, where it activates HH-target genes. The classical theory suggests that the ability of Gli proteins to be activated is controlled by interactions with upstream factors of HH signaling, including HH, PTCH1 and SMO. However, recent studies have found that Gli1 expression is in a non-canonical, SMO-independent manner (36). That is, at least part of the role of HH signaling can occur by directly activating Gli1 without upstream factors. Accordingly, research on Gli1 is a more clinically significant focus of research into the Hh signaling pathway. In this study, we identified Gli1 as an important player in breast cancer progression.
At the start of this study, differential expression of Gli1 was detected between primary cancer tissues, and their matched adjacent non-cancerous tissues. Wherein, 85% of primary cancer tissues and 25% of their matched adjacent non-cancerous tissues displayed high Gli1 expression. Data were statistically significant. In order to answer the clinical significance of this first explorative approach, we investigated a large number of samples. The analysis of 284 primary breast cancer cases revealed that Gli1 expression was strongly correlated with age and HER-2 expression. To our knowledge, youth and HER-2 overexpression were correlated with an unfavorable prognosis (37). Of patients with distant metastases, 93.3% (42/45) showed high Gli1 expression, compared to 81.2% (194/239) of patients without distant metastasis. These outcomes indicated that a high level of Gli1 expression might contribute to the invasion and progression of breast cancer. In our study, the level of Gli1 expression was a little higher than in some others (38,39); therefore the higher proportion of advanced cancer cases may have been one of the causes. Therefore, we further analyzed the relationship between Gli1 nuclear overexpression and RFS. As expected, Gli1 expression displayed a significant correlation with RFS (P=0.046). The recurrence rate was 17.8% (42/236) in the high expression (IRS=7-12) group, whereas it was only 6.2% (3/48) in the low expression (IRS=1-6) group (P=0.046). There were also significantly different outcomes between the patients with high and low levels of Gli1 expression by subset analysis. For example, when patients with T2+3+4 classifications were analyzed, the recurrence rate was 11.1% (3/27) in the low expression group and 31.2% (34/109) in the high expression group. A similar result was found in patients with N1+2+3 and clinical stage II+III, which was also shown in Kaplan-Meier curves of RFS. The above observations suggested that Gli1 could be used to predict the relapse of breast cancer.
To determine if Gli1 could serve as a clinically useful risk factor for RFS, the patients were separated according to their IRS score for Gli1 expression, and according to their clinicopathological parameters. A correlation analysis was performed, and the OR and 95% CI were estimated. After stratification, T- and N-classification, clinical stage and ER seemed to serve as risk factors (P<0.001, P<0.001, P<0.001, and P=0.002, respectively) of breast cancer relapse in the Gli1 high expression group; age was a risk factor in the low expression group (P=0.016); clinical stage was a risk factor in both groups; and PR was not a risk factor in either group (P=0.281 and 0.576, respectively). It is very important to note that the difference in Gli1 expression was related to prognosis, supporting the view that Gli1 can be used to predict relapse of breast cancer.
As metastasis can occur with the development of the tumor, some genes may constantly regulate tumor development. They may not only facilitate primary tumor initiation but also promote tumor transformation and metastasis (40). Gli1 gene is an example of this. It had been proposed that the cell-autonomous sequential increases in Gli1 activity are a crucial component of tumor progression (41,42). Consistent with this, tyr-NRASQ61K; INK4a–/– mouse melanoma showed higher levels of Gli1 in the lymph nodes after metastasis than in primary skin sites (40). Analyses of limited collections of primary and metastatic human tumors have so far been inconclusive but there is a suggestion of higher Gli1 expression in metastasis (43). In our study, 45 patients were suffering from relapses, of whom 28 have metastatic superficial nodules that had been confirmed by pathology. In order to understand the characteristics of Gli1 expression in cancer tissues, we further compared the difference in Gli1 expression in the local lymph nodes of primary carcinomas and metastatic lymph nodes of the breast cancer relapse after radical operation. Gli1 could also be detected mainly in the nuclei of leukomonocytes in the lymph nodes of breast cancer. We found 21/28 analyzed local lymph nodes exhibited increased levels of Gli1 expression compared to 27/28 in the metastatic lymph nodes. Both the proportion of samples and the score of immunoreactivity of Gli1 overexpression in the metastatic lymph nodes were higher and statistically significant (P=0.022, P=0.039, respectively). Although the proportion of positive cells and the staining intensity of Gli1 in the nucleus of the local lymph node of primary carcinoma and the metastatic lymph node of the breast cancer relapse after radical operation was not statistically significant (P=0.099), this may be due to the small sample size. There was no trend towards a difference in the staining intensity (P=0.16). The expression pattern of Gli1 indicates that Gli1 plays a prominent role in breast cancer progression and metastasis. Gli1 expression in the nuclei of leukomonocytes in the lymph nodes as part of interstitial reaction, the features and clinical significance need to be studied further and clarified.
When Gli1 expression in the cancer tissues was analyzed, it was detected in both the nuclei and cytoplasm. At the tissue level, we found Gli1 could be detected either in interstitial tissues of cancer or in cancer tissues or in both. When the primary cancer tissues of the 28 metastatic lymph nodes were taken out for analysis from the rest non-relapsed primary cancer tissues, we were surprised to find that the rate of Gli1 overexpression was 89.3% (25/28) in the relapsed group, which was much higher than of the non-relapsed group (13.4%, 32/239, P<0.001) in the interstitial tissues of primary cancer. The difference was statistically significant (P<0.001). Thus, the tissue distribution of high Gli1 expression had considerable impacts on the prognosis of breast cancer.
In short, there were interstitial molecular changes before metastasis, and high Gli1 expression in the interstitial tissue could be an indicator for breast cancer relapse. Gli1 nuclear expression measured using an immunohistochemistry-based scoring system might have a role in future multi-marker panels for human breast cancer prognosis or molecular sub-typing.
The median RFS was 25 months (range, 3-67 months) in the Gli1 high expression group in contrast to 64 months (range, 8-83 months) in the Gli1 low expression group. Our research showed that high Gli1 expression suggested that tumor recurrence occurred earlier. It was 16 months (range, 3-58 months) in contrast to 66 months of median RFS (range, 8-76 months) in interstitial tissues, which further seemed to imply early relapse after radical operation of breast cancer. The above suggested there were interstitial changes that promote metastasis in cancer tissues.
Taken together, these results indicate that Gli1 expression, together with other clinicopathological parameters, could be helpful to evaluate prognosis. Furthermore, we found that Gli1 overexpression in interstitial tissues may predict cancer relapse. Gli1 overexpression was demonstrated by both nuclear and cytoplasmic staining, in accordance with another study (13). However, multi-center studies are still required for further analysis of Gli1 expression in breast cancer tissues and the cellular and molecular mechanism of Gli1 expression.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300.
- Casey B, Hackett BP. Left-right axis malformations in man and mouse. Curr Opin Genet Dev 2000;10:257-61.
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001;15:3059-87.
- Schugar RC, Robbins PD, Deasy BM. Small molecules in stem cell self-renewal and differentiation. Gene Ther 2008;15:126-35.
- Takezaki T, Hide T, Takanaga H, et al. Essential role of the Hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci 2011;102:1306-12.
- Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature 2003;422:313-7.
- Karhadkar SS, Bova GS, Abdallah N, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature 2004;431:707-12.
- El-Zaatari M, Tobias A, Grabowska AM, et al. De-regulation of the sonic hedgehog pathway in the InsGas mouse model of gastric carcinogenesis. Br J Cancer 2007;96:1855-61.
- Cui W, Wang LH, Wen YY, et al. Expression and regulation mechanisms of Sonic Hedgehog in breast cancer. Cancer Sci 2010;101:927-33.
- Souzaki M, Kubo M, Kai M, et al. Hedgehog signaling pathway mediates the progression of non-invasive breast cancer to invasive breast cancer. Cancer Sci 2011;102:373-81.
- Kasper M, Regl G, Frischauf AM, et al. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer 2006;42:437-45.
- Borgquist S, Holm C, Stendahl M, et al. Oestrogen receptors alpha and beta show different associations to clinicopathological parameters and their co-expression might predict a better response to endocrine treatment in breast cancer. J Clin Pathol 2008;61:197-203.
- ten Haaf A, Bektas N, von Serenyi S, et al. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer 2009;9:298.
- Rydén L, Jirström K, Bendahl PO, et al. Tumor-specific expression of vascular endothelial growth factor receptor 2 but not vascular endothelial growth factor or human epidermal growth factor receptor 2 is associated with impaired response to adjuvant tamoxifen in premenopausal breast cancer. J Clin Oncol 2005;23:4695-704.
- Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989;63:181-7.
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108.
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature 1980;287:795-801.
- Echelard Y, Epstein DJ, St-Jacques B, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 1993;75:1417-30.
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 1993;75:1431-44.
- Riddle RD, Johnson RL, Laufer E, et al. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 1993;75:1401-16.
- Chang DT, López A, von Kessler DP, et al. Products, genetic linkage and limb patterning activity of a murine hedgehog gene. Development 1994;120:3339-53.
- Hatsell S, Frost AR. Hedgehog signaling in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia 2007;12:163-73.
- Bak M, Hansen C, Friis Henriksen K, et al. The human hedgehog-interacting protein gene: structure and chromosome mapping to 4q31.21-->q31.3. Cytogenet Cell Genet 2001;92:300-3.
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov 2006;5:1026-33.
- Sasaki H, Nishizaki Y, Hui C, et al. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 1999;126:3915-24.
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell 2004;6:103-15.
- Buttitta L, Mo R, Hui CC, et al. Interplays of Gli2 and Gli3 and their requirement in mediating Shh-dependent sclerotome induction. Development 2003;130:6233-43.
- Dai P, Akimaru H, Tanaka Y, et al. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem 1999;274:8143-52.
- Ikram MS, Neill GW, Regl G, et al. GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J Invest Dermatol 2004;122:1503-9.
- Koyabu Y, Nakata K, Mizugishi K, et al. Physical and functional interactions between Zic and Gli proteins. J Biol Chem 2001;276:6889-92.
- Mao J, Maye P, Kogerman P, et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J Biol Chem 2002;277:35156-61.
- Kubo M, Nakamura M, Tasaki A, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res 2004;64:6071-4.
- Yanai K, Nagai S, Wada J, et al. Hedgehog signaling pathway is a possible therapeutic target for gastric cancer. J Surg Oncol 2007;95:55-62.
- Kasai K, Inaguma S, Yoneyama A, et al. SCL/TAL1 interrupting locus derepresses GLI1 from the negative control of suppressor-of-fused in pancreatic cancer cell. Cancer Res 2008;68:7723-9.
- Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J 2009;28:663-76.
- Rahnama F, Shimokawa T, Lauth M, et al. Inhibition of GLI1 gene activation by Patched1. Biochem J 2006;394:19-26.
- Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med 2009;360:790-800.
- O’Toole SA, Machalek DA, Shearer RF, et al. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res 2011;71:4002-14.
- Souzaki M, Kubo M, Kai M, et al. Hedgehog signaling pathway mediates the progression of non-invasive breast cancer to invasive breast cancer. Cancer Sci 2011;102:373-81.
- Bernards R, Weinberg RA. A progression puzzle. Nature 2002;418:823.
- Sanchez P, Hernández AM, Stecca B, et al. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci U S A 2004;101:12561-6.
- Stecca B, Mas C, Clement V, et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A 2007;104:5895-900.
- Feldmann G, Dhara S, Fendrich V, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res 2007;67:2187-96.


