microRNAs and ceRNAs: RNA networks in pathogenesis of cancer
Introduction
microRNAs (miRNAs) are small, evolutionarily conserved, single-stranded RNAs of 18-25 nucleotides in length that play major roles in gene regulation. miRNAs were shown to inhibit their target genes through binding to miRNA response elements (MREs) on the 3' untranslated regions (UTRs) of target RNA transcripts with imperfect complementarity, and leading to decreased expression of their target proteins either by mRNA degradation or translational inhibition (1). Single mRNA usually contains MREs for multiple miRNAs. At the same time, individual miRNA often targets up to 200 transcripts which are diverse in their function. miRNAs have been shown to suppress the expression of important cancer-related genes and have been proved useful in the diagnosis and treatment of cancer (2). However, the mechanism by which the miRNA could be regulated is largely unknown.
Based on the fact that the MREs on the RNA transcripts could be predicted and validated by many types of software and experimental techniques, Salmena et al. hypothesized that all types of RNA transcripts talk to each other by miRNA-mediated language. The previous pattern, “miRNAs→RNAs”, could be replaced by “RNAs→miRNAs→RNAs”. RNA transcripts sharing multiple MREs in their 3' UTRs communicate to each other and regulate the expression levels by competing for a limited pool of miRNAs. The upregulation of a given mRNA leads to the increased total number of MREs, which exceeds their targeting miRNAs. As a result, the targeting miRNAs would be diluted with the derepression of other mRNAs sharing the same MREs. RNAs involved in this process are known as competitive endogenous RNAs (ceRNAs). The large scale identification of ceRNAs constructs ceRNAs regulatory networks (Figure 1) (3,4).
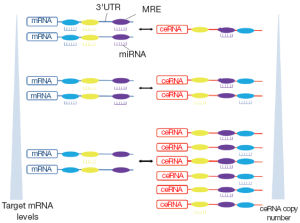
In this system, in addition to conventional function of RNAs to translate to proteins, RNAs may exert another function through their ability to regulate other RNAs (5-8). Furthermore, general studies in gene function usually focus on gene coding regions by overexpression or knocking down of the coding sequence (CDS). The function of UTRs in transcript will be neglected. However, hypothesis of ceRNAs network provides a new understanding on the regulatory function of full transcript including 3' UTR and CDS. The discovery of ceRNAs not only dictates a reevaluation of our understanding of gene regulatory networks but also opens up the possibility of a new biological mechanism that could be targeted by oligonucleotide gene therapy (3).
Methods of Identification of ceRNAs
The identification of ceRNAs depends on the precision of MREs prediction on RNAs, as well as miRNA target prediction (4). miRNAs target prediction algorithms and experimental approaches make it possible to identify MREs related to the RNA-induced silencing complex (RISC). Tay et al. devised a novel approach termed mutually targeted MRE enrichment (MuTaME) to predict the candidate ceRNAs for distinct target. The possibility of putative ceRNAs rank on MuTaME scores generated from the rna22 miRNA target prediction algorithm, which is mainly based on the number of miRNAs with which ceRNAs and target transcripts shared, as well as the distribution of MREs in both ceRNAs and target transcripts (9). Sumazin et al. presented Hermes, a new multivariate analysis method, to analyze candidates according to genome-wide expression profiles of mRNAs and miRNAs from the same tumor samples (10). On the other hand, the novel biochemical techniques, including the argonaute high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (Ago HITS-CLIP) and photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP), provide general platforms for identification of ceRNAs, due to restricting the precise sequences for miRNA-mRNA interactions (11).
Condition that ceRNA network may occur
ceRNAs network contains transcripts that share multiple MREs targeted by multiple miRNAs. Thus, whether ceRNAs network has its function largely depends on the concentration of the ceRNAs and their miRNAs. Changes in the ceRNA level need to be large enough to effectively compete for miRNAs binding. Appropriate miRNA level is another important factor impacting ceRNAs network, since there is a balance between ceRNAs and target RNAs for the distribution of miRNAs. In addition, ceRNA networks would also rely on the subcellular distribution and tissue specificity of RNAs and miRNAs existent in a proper cell type at a proper moment (4).
ceRNA association with cancer and its classification
Cancers develop from the accumulation of genomic mutations and epigenetic changes that alter gene function and expression. Accordingly, DNA-based somatic base-pair mutations, changes in DNA copy number, chromosomal translocation, as well as mRNA-based gene fusions, alternative splicing are commonly observed in cancer (12). As a result, such events inducing altered expression of UTRs in transcripts would influence MRE levels, or introduce new MREs into the cells. Changes in MREs of ceRNAs impact the capacity of a proper mRNA transcript to attach or titrate miRNAs. Consequently, the perturbed ceRNA network may contribute to diseases and cancers (4,9,10,13,14). There are three types of transcriptome belonging to ceRNA: protein coding genes, transcribed pseudogenes, and long noncoding RNAs.
Protein coding genes
The study by Tay et al. identified a series of endogenous protein-coding transcripts, serine incorporator 1 (SERINC1), vesicle-associated membrane protein associated protein A (VAPA), CCR4-NOT transcription complex, subunit 6-like (CNOT6L), as phosphatase and tensin homolog (PTEN) ceRNAs which regulate PTEN levels in a miRNA-dependent manner. Due to the same target miRNAs that PTEN and ceRNAs shared, deletion of ceRNA by siRNAs facilitated more miRNAs contacting with PTEN 3' UTR, and resulted in a significant reduction in PTEN protein levels. Moreover, mutual reciprocal regulation of transcripts in the PTEN ceRNA network is validated by the fact not only that PTEN downregulation modulates ceRNA expression, but also that PTEN ceRNAs are coexpressed with PTEN in human samples (9). Further studies by the same group isolated zinc finger E-box binding homeobox 2 (ZEB2) as a PTEN ceRNA by a Sleeping Beauty insertional mutagenesis screen in oncogenic BRAF-induced mouse model of melanoma. Consistent with other PTEN ceRNAs, ZEB2 levels were commonly reduced and significantly correlated with PTEN in a set of human cancers (13). Recently, it has been reported that 3' UTR of versican could serve as a ceRNA in up-regulating the expression of versican, CD34, and fibronectin, leading to the development of hepatocellular carcinoma by controlling the miRNA activity (15). Furthermore, based on Hermes, a novel bioinformatics multivariate analysis method, Sumazin et al. predicted an extensive miRNA-mediated ceRNA network, including about 7,000 genes, which regulate established oncogenic pathways in glioblastoma (10).
Transcribed pseudogenes
Although pseudogenes share high sequence identity with their ancestral protein coding genes, they are lack of protein-coding ability, due to various genetic disablements including premature stop codes, frameshifts, insertions, or deletions (16). Nevertheless, sequencing efforts have revealed nearly 19,000 pseudogenes in human genome, many of which are transcribed and are often well conserved, suggesting that selective pressure to maintain pseudogenes exists (17).
Previous studies focus on miRNA dependent regulation, also known as “miRNAs→RNAs”. However, the studies by Seitz et al. hypothesized that most miRNA targets regulate the miRNA. Since there is a high homology between pseudogenes and their protein coding partners, numerous miRNA targets identified by computational prediction, especially the transcribed pseudogenes, may actually be competitive inhibitors of miRNA function, preventing miRNAs from binding to their authentic targets by sequestering and titrating them (18). Recently, the research by Poliseno et al. also supported the similar theory that pseudogenes can regulate miRNAs, and influence miRNAs’ availability for other RNAs. They discovered that PTENP1, the pseudogene of tumor suppressor PTEN sharing high sequence homology with PTEN, can compete with PTEN for the same pool of miRNAs through many conserved MREs. As a result, PTENP1 3' UTR overexpression increased both PTEN transcript and protein in a DICER-dependent manner. Moreover, the correlation between PTENP1 and PTEN in normal human tissues, and prostate tumor samples, as well as the direct association between PTENP1 copy number and PTEN expression in colon cancer tissues suggests that PTENP1 transcript levels can regulate PTEN expression, and act as a tumor suppressor gene (19). KRAS and its pseudogene KRAS1P have the similar trend to PTEN and PTENP1. KRAS1P 3' UTR overexpression induced KRAS mRNA abundance. The transcript levels of KRAS and KRAS1P are directly correlated in prostate cancer, neuroblastoma, retinoblastoma and hepatocellular carcimoma, which indicates KRAS1P plays a proto-oncogenic role in cancer and acts as a ceRNA of KRAS for the same pool of miRNAs. Sequence alignment also uncovered that a miRNA binding site, MRE, is conserved in gene and its pseudogene counterpart, as OCT4 and OCT4-pg1-5, FOXO3 and FOXO3B, or E2F3 and E2F3P1 (19).
Long noncoding RNAs
With the expanding of the number of long noncoding RNAs (lncRNAs), more and more studies pay attention to the role of lncRNAs in diseases. Especially, a subset of lncRNAs is associated with epigenetic mechanisms (20,21). Through the Ago HITS-CLIP technique, a recent Argonaute (Ago)-bound transcripts analysis discovered that lncRNAs are the targets of miRNAs (11,22). Highly up-regulated in liver cancer (HULC) is one of lncRNAs, and plays an important role in tumorigenesis. HULC acts as a ceRNA, which down-regulates a series of miRNAs activities, including miR-372. As a result, the translational level of miR-372 target gene, PRKACB, was derepressed in liver cancer (23). Since noncoding RNAs are not involved in active protein translation process, they are more effective ceRNAs than protein-coding RNAs in miRNAs binding (24).
ceRNAs and cancer therapy
The perturbations of ceRNA networks could lead to carcinogenesis and other diseases (4,9,10,13,14). However, it provides a new perspective to explain disease processes and offer new opportunities to manipulate ceRNA networks through miRNA competition for cancer therapy. Although the mechanism of ceRNAs needs largely to be explored, the techniques are available to identify MREs on the transcripts, and recognize the related binding miRNAs as the ceRNA language.
With regard to the therapies against miRNA function, many studies found that “miRNA sponges”, the artificial miRNA decoys, which are oligonucleotide constructs with multiple copies of the same MRE in tandem targeting for only one miRNA, have already been used as a RNA-based therapy to deplete individual miRNA in cells and transgenic animals (25-30). In contrast, ceRNAs are “endogenous sponges” which are able to regulate the distribution of miRNAs on their natural targets. Unlike artificial miRNA sponges, ceRNAs contain MREs to combine different miRNAs. Therefore, they could influence the multiple targets containing multiple miRNAs. The application of ceRNA sponges could be an ideal approach for developing therapies against miRNA function (4). Although both miRNA sponges and ceRNA sponges provide strategies for miRNA loss-of-function studies, more study is required to rule out off-target effects and evaluate their potential effects.
Taken together, the identification of the ceRNA mechanism expands the theory of the dynamics and complexity of the miRNA regulatory network and provides more challenge in the development of new strategies for miRNA-based cancer diagnosis and therapy.
Acknowledgements
This study was supported by the grants from the National Natural Science Foundation of China (No. 81272766), Capital Medical Development and Research Foundation (No. 2009-2093), Clinical Characteristics and Application Research of Capital (No. Z121107001012130), Beijing Natural Science Foundation (No. 7132054), and New Star of Science and Technology Program of Beijing (No. 2011060).
Disclosure: The authors declare no conflict of interest.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33.
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69.
- Khvorova A, Wolfson A. New competition in RNA regulation. Nat Biotechnol 2012;30:58-9.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8.
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011;39:D152-7.
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105.
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004.
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9.
- Tay Y, Kats L, Salmena L, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011;147:344-57.
- Sumazin P, Yang X, Chiu HS, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 2011;147:370-81.
- Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol 2010;17:1169-74.
- Berger MF, Levin JZ, Vijayendran K, et al. Integrative analysis of the melanoma transcriptome. Genome Res 2010;20:413-27.
- Karreth FA, Tay Y, Perna D, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011;147:382-95.
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358-69.
- Fang L, Du WW, Yang X, et al. Versican 3'-untranslated region (3'-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J 2013;27:907-19.
- D'Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief Funct Genomic Proteomic 2004;3:157-67.
- Pink RC, Wicks K, Caley DP, et al. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA 2011;17:792-8.
- Seitz H. Redefining microRNA targets. Curr Biol 2009;19:870-3.
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8.
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223-7.
- Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009;106:11667-72.
- Chi SW, Zang JB, Mele A, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009;460:479-86.
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83.
- Gu S, Jin L, Zhang F, et al. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol 2009;16:144-50.
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA 2010;16:2043-50.
- Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods 2009;6:63-6.
- Brown BD, Gentner B, Cantore A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 2007;25:1457-67.
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012;482:347-55.
- Loya CM, Lu CS, Van Vactor D, et al. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods 2009;6:897-903.
- Zhu Q, Sun W, Okano K, et al. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J Biol Chem 2011;286:31749-60.


