DNA repair gene XRCC1 polymorphisms and susceptibility to childhood acute lymphoblastic leukemia: a meta-analysis
Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignant tumor in the children which comprises over 80% of all the acute leukemia. It is estimated that approximately 33.6 in every 1,000,000 children under 15 years old will develop ALL (1). The etiology and pathogenesis of ALL are unclear up to now. Previous studies make a plausible hypothesis that crucial sequential events involving specific chromosomal translocations or fusion genes as the first hits to initiate ALL and further genetic or epigenetic events such as gene deletions or mutations as the second hits to cooperate to the outbreak of ALL (2-5). One of the most important initiating events is thought to result from the misrepair of double-strand DNA breaks (DSB) during non- homologous end-joining (NHEJ) (6-8). Considering roles in repairing of DSB, X-ray repair cross-complementing group 1 (XRCC1) gene is one of the most important DNA repair genes (9,10). XRCC1 also participates in single-strand DNA break (SSB) repair pathway for the repair of DNA destruction which occurs very frequently in mammals and base excision repair (BER) pathway which operates on small lesions caused normally by endogenous substances or xenobiotics. Moreover, it is reported that DNA repair function could be modified by genetic polymorphisms (11). It could cause genetic instability and even carcinogenesis if DNA repair capacity is of deficiency (12,13). Also reported by relevent studies, the disruption of XRCC1 in mice results in early embryonic lethality in the BER pathway (14,15), and an excess of deletions found among induced mutations in EM-C11 (one cell line identified as defective in XRCC1 function) perhaps result from reduced ligation efficiency of SSB (16). Therefore, the size of DNA repair capacity modified by XRCC1 gene polymorphisms makes different hereditary susceptibility to ALL in different population. In other words, XRCC1 gene polymorphisms may be associated with childhood ALL. There are three common single nucleotide polymorphism (SNP) sites in XRCC1 gene encoding region, which are codon 399 (extron 10, G→A, Arg→Gln), codon 194 (extron 6, C→T, Arg→Trp), and codon 280 (extron 9, G→A, Arg→His) respectively. Molecular epidemiological studies on the association between genetic predisposition of children ALL and XRCC1 polymorphisms have presented some contradictory results (17-24). In this paper, we performed a quantitative synthesis by meta-analysis method to evaluate the size of the association between XRCC1 polymorphisms and children ALL.
Materials and methods
Search strategy
All the studies were identified by a computerized literature search of the PubMed, EMBASE, Elsevier Science Direct, ProQuest and SpringerLink with the keywords of “XRCC1” or “X-ray repair cross-complementing group 1” or “DNA repair gene”, “polymorphism” or “variant” or “Arg399Gln, Arg194Trp, Arg280His”, “acute lympho* leukemia”, “childhood” or “pediatric”. We also searched the database of Chinese National Knowledge Infrastructure (CNKI), Halis, and ChinaInfo using “ALL” and “genetic polymorphism” as the keywords. All finished prior to Dec 1st, 2010 in any language. References of the retrieved publications were also screened.
Inclusion criteria
The inclusion criteria comprise: (I) only the case-control studies on the association between XRCC1 polymorphisms and the risk of children ALL were considered; (II) at least one of the XRCC1 polymorphisms, Arg399Gln, Arg194Trp and Arg280His, was included; (III) essential information about the distribution on genotype frequency should be described in details, and the data should be extractable; (IV) there were detailed descriptions on study subject and DNA sources; and (V) the authors must offer sample size, odds ratios (OR) and 95% confidence interval (95% CI).
Exclusion criteria
The exclusion criteria comprise: (I) review or abstract; (II) only with case or non-case-control studies; (III) adult ALL or adult and childhood ALL; (IV) no relationship between studies and ALL; and (V) without essential information. Additionally, we selected the latest one when some literatures were repeated.
Data extraction
Two investigators independently extracted and checked the information, and discussed with each other to reach a consensus. The data included author, journal, the year of publication, country of origin, race, demographic characteristics, sample size of the cases and controls, genotyping information, OR, 95% CI, and matching factors.
Quality score assessment
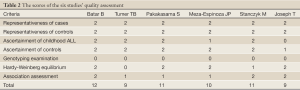
The same two reviewers independently assessed the quality of studies by quality assessment scores which were modified from previous meta-analyses of molecular association studies (25-27) (Table 1). And the scores were based on both traditional epidemiologic considerations and genetic issues (28,29). Total scores ranged from 9 to 12 (Table 2).

Full Table
Full Table
Statistical analysis
A Q-statistic test was performed to analyze the heterogeneity of the included literatures. And we set the significance level at 0.1 (30). If P≥0.1, which means there was no heterogeneity between the individual studies, a fixed-effects model (Peto Mantel-Haenszel) would be used to pool the results, otherwise, a random-effects model (DerSimonian and Laird) was used (31). All of the statistical analyses were performed with Review Manager 4.2 to show the combined results by drawing forest plot.
We used the meta-analysis method to evaluate the relationship between the risk of childhood ALL and the Arg399Gln, Arg194Trp and Arg280His polymorphisms. OR and 95% CI were chosen as the effect magnitude and significance level as α=0.05. Statistical tests were two-sided test. For Arg399Gln, we compared allele Gln with allele Arg on the risk of developing children ALL. Arg/Arg wild genotype was selected as reference to compare the risk of children ALL patients with homozygote Gln/Gln genotype and heterozygote Arg/Gln genotype. Supposing the mutated gene was the dominant gene, the OR of the dominant model (Arg/Gln+Gln/Gln vs. Arg/Arg) and the recessive model (Gln/Gln vs. Arg/Gln+Arg/Arg) was calculated. The same method was applicable to Arg194Trp and Arg280His. At the same time, we also did the subgroup analysis and the sensitivity analysis and provided publication bias estimate with funnel plot.
Results
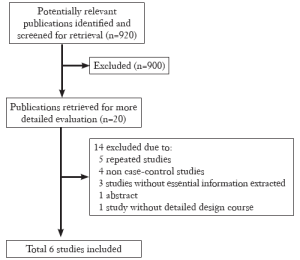
Eligible studies
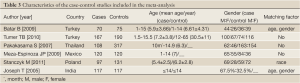
According to the search criteria, there were total 6 English studies (17-24) including 679 cases and 950 controls accepted for our meta-analysis by screening among the 20 candidate literatures (Figure 1). Five articles were case-control studies based on healthy people, the rest one based on hospital people (23), all of whom from the six literatures came from different races, but the cases and controls in each study were from the same ethnic group. All patients of the total studies were less than sixteen years old while controls from different researches had different age ranges excepted that two studies had no description about the age of controls (19,22). The sex ratio varied in each literature. Three articles were matched by age, gender or race (17,20,23). Hardy-Weinberg equilibrium (HWE) test was made in five articles (16-18,20,21). All the studies used polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method to identify XRCC1 polymorphism genotype. Genomic DNA was extracted from the venous blood of patients, who were in remission, therapy or diagnosis. There were five researches having illustration on the diagnosis of childhood ALL (17-22), and only Tumer TB et al. made the risk stratification of children ALL in proportion. Table 3 describes characteristics of included literatures. Tables 4,5,6 describe the distribution of XRCC1 Arg399Gln, Arg194Trp, Arg280His genotype in childhood ALL cases and controls.
Full Table
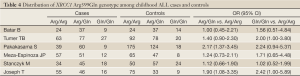
Full Table
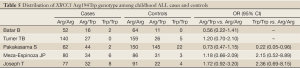
Full Table

Full Table
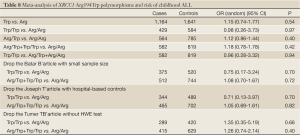
Quantitative synthesis
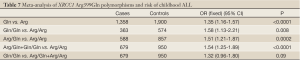
Six studies including 679 cases and 950 controls on relationship between XRCC1 Arg399Gln polymorphism and children ALL were accepted for our meta-analysis, five articles including 582 cases and 819 controls on the XRCC1 Arg194Trp polymorphism of the developing risk of children ALL were included, and three literatures including 345 cases and 554 controls on the XRCC1 Arg280His polymorphism of the developing risk of children ALL were included. All the DNA samples were from peripheral venous blood. And the cases’ DNA samples were extracted from different stages including primary diagnosis, treatment and complete remission. Only Stanczyk M et al. had DNA duplicate detection and none had blind method on XRCC1 genotyping. Tumer TB and Stanczyk M also discussed the association of XRCC1 Arg399Gln polymorphism and together with other genes polymorphisms with the developing risk of children ALL.
Arg399Gln
No significant between-study heterogeneities were found concerning the relationship between XRCC1 Arg399Gln polymorphism and children ALL with P>0.1. Hence, we used a fixed-effects mode to pool data. The risk of ALL for children with Gln allele was 1.35 times to that with Arg allele, which had statistically significant difference (P<0.0001). Compared to Arg/Arg wild genotype, both of the homozygote Gln/Gln genotype and heterozygote Arg/Gln genotype could increase the risk of children ALL. (Gln/Gln vs. Arg/Arg: OR, 1.58; 95% CI, 1.13-2.21; P=0.008) (Arg/Gln vs. Arg/Arg: OR, 1.51; 95% CI, 1.21-1.87; P=0.0002). Similarly, the Arg399Gln dominant model was also associated with childhood ALL (Arg/Gln+Gln/Gln vs. Arg/Arg: OR, 1.54; 95% CI, 1.25-1.89; P<0.0001). However, no associations were found in the recessive model (Gln/Gln vs. Arg/Gln+Arg/Arg: OR, 1.32; 95% CI, 0.96-1.80; P=0.09) (Table 7). From the forest plot we can see that the Gln allele in all five ethnic groups was prone to be associated with childhood ALL just with different degrees of correlation, and Thai race and Indian race had statistically significant differences (OR =1.67, 95% CI, 1.20-2.33, P=0.002 in Thai race; OR =1.79, 95% CI, 1.19-2.71, P=0.005 in Indian race). The overall result (OR =1.35, 95% CI, 1.16-1.57, P<0.0001) suggested a relationship between Gln allele and childhood ALL in all different ethnic groups described above (Figure 2).
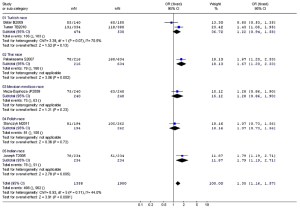
Full Table
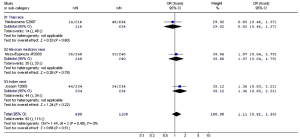
Arg194Trp
The studies about the relationship between XRCC1 Arg194Trp and childhood ALL were heterogeneous (except the comparison between Arg/Trp and Arg/Arg using the fixed-effects model, P=0.17). Therefore, we used the random-effects model to pool data showing no association between Arg194Trp polymorphism and childhood ALL (Table 8). The ethnic subgroup analysis demonstrated that the relationship between Trp allele and childhood ALL in the four ethnic groups was variant. As for Thai race, it represented a protected role for Trp allele in the childhood ALL development (OR =0.67, 95% CI, 0.47-0.97, P=0.03). While as for Indian race, it may increase the risk of childhood ALL with Trp allele (OR =1.75, 95% CI, 1.07-2.89, P=0.03). The rest two ethnic groups with Trp allele provided no relationship with childhood ALL. The synthesized result revealed no relationship between Trp allele and childhood ALL in the four ethnic groups (OR =1.15, 95% CI, 0.74-1.77, P=0.54) (Figure 3).
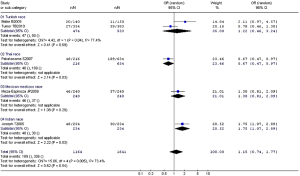
Full Table
Arg280His
According to the meta-analysis, there was no relationship between XRCC1 Arg280His and childhood ALL with no significant between-study heterogeneities using the fixed-effects model to pool data (Table 9). Ethnic group analysis showed no relationship between His allele and childhood ALL within each race (Figure 4).
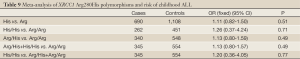
Full Table
Sensitivity analysis and publication bias evaluation
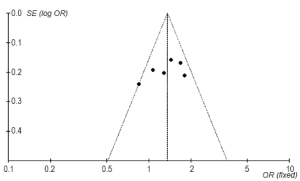
There were no significant changes in conclusions about the association between Arg194Trp polymorphism and childhood ALL before and after excluding some articles (Table 8), which might be the sources of heterogeneity. Additionally, we analyzed the publication bias by drawing funnel plot, from which we can see that six points were approximately symmetrical in the inverted funnel plot (Figure 5).
Discussion
Human genome DNA is composed of approximately 3 billion base pairs, which are frequently influenced by endogenous metabolite, medical medicine and massive environment mutagen resulting in the damage of DNA integrity in the physiological activity. If this damage can not be recognized and repaired in time, the error replication, gene mutation and even tumor may occur. XRCC1 gene product is one of the important proteins participated in BER, SSB and DSB repair pathways and the gene is located at 19q13.2-13.3. It is reported that XRCC1 gene knockout mice may die in the early embryogenesis (14) and the cells with this XRCC1 gene defection are very sensitive to ionizing radiation and alkylating agent (9), which illustrated that XRCC1 gene plays an important role in the DNA integrity maintaining and individual development. XRCC1 gene has three common SNPs, codon 399, 194 and 280, which maybe change the repair ability of DNA. These genetic susceptibility genes are considered as potential hazard factors for childhood ALL (32).
Meta-analysis is a method through collecting comparable literatures published or unpublished, and applying certain statistical method to synthesize the independent results of studies with the same research target to obtain a combined quantitative conclusion, or a reason why similar studies will have different results, which will be strongly scientific, repeatable and objective.
Based on our meta-analysis about the association between XRCC1 SNPs, Arg399Gln, Arg194Trp and Arg280His, and childhood ALL, we found that the Arg399Gln SNP possibly increased the children ALL risk with no between-study heterogeneity. On the contrary, both of XRCC1 Arg194Trp and Arg280His polymorphisms did not contribute to childhood ALL. But the Arg399Gln SNP might have small absolute risk effect for the occurrence of childhood ALL on the basis of low OR ranging from 1.32 to 1.58, which was consistent with previous genetic susceptibility researches (33-35). Additionally, some literatures reported that folate deficiency can contribute to an elevation in DNA breaks (36) and the successful repair of DNA breaks during fetal hematopoiesis can be modified by genetic and dietary factors in the folate pathway (37). Therefore, it is necessary to research the interaction between XRCC1 repair ability and folate metabolism for the contribution to fetal ALL.
Epidemiologic data show that ethnic and racial groups differ significantly in terms of cancer incidence and mortality rates (38). On this point, we also achieve some discoveries in this study. Our ethnic subgroup analysis manifested that XRCC1 Arg399Gln SNP did some contributions to children with ALL in different ethnics. It seems to be a dangerous factor for childhood ALL in each race as all the ORs were above 1.0. Especially in Asia countries, such as Thailand and India, the statistical data of which had significant difference. However, the correlation between childhood ALL and Arg194Trp SNP was not the same in each ethnic. In Thai race, Arg194Trp SNP was a protective factor while in India it is a risk factor and no relationship was found in the rest ethnic groups. For Arg280His SNP, ethnic group analysis showed no association between His allele and childhood ALL within each race. The racial or ethnic variation in cancer risk may reflect differences in environmental exposures or socio-economic and demographic factors as well as inherent biological susceptibility (39). Consequently, careful attention must be paid to racial stratification when we research the relationship between gene polymorphisms and disease.
Studies about the association between XRCC1 Arg194Trp polymorphism and childhood ALL were heterogeneous. Sensitivity analysis was conducted by excluding the relatively poor quality of literatures, but results hardly changed. It demonstrated good stability of the results, but no heterogeneous sources were found. However, perhaps racial differences are sources of heterogeneity which we have had a very detailed discussion. Additionally, publication bias assessment analysis displayed that six points in the funnel plot are basically symmetrical, indicating less impact on the results. But the conclusion should be considered preliminary because of the small number of studies included.
In our article, there are still some limitations. First, there was one study based on hospital population, which was difficult to determine whether genetic polymorphism was related to ALL or not. Perhaps this polymorphism was associated with other disease rather than ALL. Second, only three articles matched confounding factors such as gender, age and race in the case and control group. Thus results of the rest researches might be disturbed by the unmatched confounding factors. Third, only one article did not abide by HWE among the six included studies except an article without HWE inspection, which suggested that it was wrong in genotyping or inappropriate in choosing control group (40). Forth, the overall sample sizes of included literatures are quite small. Fifth, some valuable studies are probably omitted by the impact of publication bias, because articles with positive results or in English are easier to be published. So we should have larger sample size, rigorous design approach, perfect retrieval strategy and reasonable inclusion and exclusion criteria in the future studies.
Although there were some deficiencies in this article, the comprehensive research results showed that XRCC1 Arg399Gln polymorphism could increase the risk of childhood ALL, which was to be significant to some extent. Because this not only can deeply enhance our knowledge of the childhood ALL pathogenesis, which will offer potential targets for therapeutic intervention, but also assess the risk of childhood ALL in order to take effective measures to protect vulnerable populations. Meanwhile, it is essential to study the correlation between ALL and gene polymorphism in Chinese children. More researches are needed to investigate the relationship between DNA repair genes polymorphisms and childhood ALL.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Silverberg E. Cancer statistics, 1979. CA Cancer J Clin 1979;29:6-21. [PubMed]
- Ziegelberger G, Baum C, Borkhardt A, et al. Research recommendations toward a better understanding of the causes of childhood leukemia. Blood Cancer J 2011;1:e1.
- Rudant J, Amigou A, Orsi L, et al. Fertility treatments, congenital malformations, fetal loss, and childhood acute leukemia: The ESCALE study (SFCE). Pediatr Blood Cancer 2013;60:301-8. [PubMed]
- Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer 2006;6:193-203. [PubMed]
- Rossig C, Juergens H. Aetiology of childhood acute leukaemias: current status of knowledge. Radiat Prot Dosimetry 2008;132:114-8. [PubMed]
- Kim AS, Eastmond DA, Preston RJ. Childhood acute lymphocytic leukemia and perspectives on risk assessment of early-lifestage exposures. Mutat Res 2006;613:138-60. [PubMed]
- Hassanzadeh J, Mohammadi R, Rajaeefard AR, et al. Maternal and prenatal risk factors for childhood leukemia in southern of iran. Iran Red Crescent Med J 2011;13:398-403. [PubMed]
- Emerenciano M, Koifman S, Pombo-de-Oliveira MS. Acute leukemia in early childhood. Braz J Med Biol Res 2007;40:749-60. [PubMed]
- Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955-69. [PubMed]
- Lee Y, Katyal S, Li Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci 2009;12:973-80. [PubMed]
- Lee JM, Lee YC, Chen CJ, et al. Application of genetic polymorphisms in DNA repair in the prediction of cancer susceptibility and clinical outcome. Bentham Science Publishers 2003;1:175-86.
- Goode EL, Ulrich MC, Potter DJ, et al. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 2002;11:1513-30. [PubMed]
- Jiang J, Zhang X, Yang H, et al. Polymorphisms of DNA repair genes: ADPRT, XRCC1, and XPD and cancer risk in genetic epidemiology. Methods Mol Biol 2009;471:305-33. [PubMed]
- Tebbs RS, Flannery ML, Meneses JJ, et al. Requirement for the XRCC1 DNA base excision repair gene during early mouse development. Dev Biol 1999;208:513-29. [PubMed]
- Horton JK, Watson M, Stefanick DF, et al. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res 2008;18:48-63. [PubMed]
- Op het Veld CW, Jansen J, Zdzienicka MZ, et al. Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CHO9 and its xrcc1-deficient derivative EM-C11. Mutat Res 1998;398:83-92. [PubMed]
- Batar B, Guven M, Baris S, et al. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res 2009;33:759-63. [PubMed]
- da Silva Silveira V, Canalle R, Scrideli CA, et al. Polymorphisms of xenobiotic metabolizing enzymes and DNA repair genes and outcome in childhood acute lymphoblastic leukemia. Leuk Res 2009;33:898-901. [PubMed]
- Meza-Espinoza JP, Peralta-Leal V, Gutierrez-Angulo M, et al. XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia. Genet Mol Res 2009;8:1451-8. [PubMed]
- Stanczyk M, Sliwinski T, Cuchra M, et al. The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia. Mol Biol Rep 2011;38:445-51. [PubMed]
- Tumer TB, Yilmaz D, Tanrikut C, et al. DNA repair XRCC1 Arg399Gln polymorphism alone, and in combination with CYP2E1 polymorphisms significantly contribute to the risk of development of childhood acute lymphoblastic leukemia. Leuk Res 2010;34:1275-81. [PubMed]
- Pakakasama S, Sirirat T, Kanchanachumpol S, et al. Genetic polymorphisms and haplotypes of DNA repair genes in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer 2007;48:16-20. [PubMed]
- Joseph T, Kusumakumary P, Chacko P, et al. DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia. Cancer Lett 2005;217:17-24. [PubMed]
- Canalle R, Silveira VS, Scrideli CA, et al. Impact of thymidylate synthase promoter and DNA repair gene polymorphisms on susceptibility to childhood acute lymphoblastic leukemia. Leuk Lymphoma 2011;52:1118-26. [PubMed]
- Thakkinstian A, McEvoy M, Minelli C, et al. Systematic review and Meta-analysis of the association between β2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol 2005;162:201-11. [PubMed]
- Gao J, Shan G, Sun B, et al. Association between polymorphism of tumor necrosis factor alpha-308 gene promoter and asthma: a meta-analysis. Thorax 2006;61:466-71. [PubMed]
- Thakkinstian A, D’Este C, Eisman J, et al. Meta-analysis of molecular association studies: vitamin D receptor gene polymorphisms and BMD as a case study. J Bone Miner Res 2004;19:419-28. [PubMed]
- Attia J, Thakkinstian A, D’Este C. Systematic review of Meta-analyses of molecular association studies: methodologic lesson for genetic epidemiology. J Clin Epidemiol 2003;56:297-303. [PubMed]
- Boccia S, De Feo E, Gallì P, et al. A systematic review evaluating the methodological aspects of meta-analyses of genetic association studies in cancer research. Eur J Epidemiol 2010;25:765-75. [PubMed]
- Fleiss JL. Statistical methods for rates and proportions. 2nd ed. New York: John Wiley, 1981:161-5.
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820-6. [PubMed]
- Karathanasis NV, Choumerianou DM, Kalmanti M. Gene Polymorphisms in Childhood ALL. Pediatr Blood Cancer 2009;52:318-23. [PubMed]
- Sherborne AL, Hemminki K, Kumar R, et al. Rationale for an international consortium to study inherited genetic susceptibility to childhood acute lymphoblastic leukemia. Haematologica 2011;96:1049-54. [PubMed]
- Vijayakrishnan J, Houlston RS. Candidate gene association studies and risk of childhood acute lymphoblastic leukemia: a systematic review and meta-analysis. Haematologica 2010;95:1405-14. [PubMed]
- Chokkalingam AP, Buffler PA. Genetic susceptibility to childhood leukaemia. Radiat Prot Dosimetry 2008;132:119-29. [PubMed]
- Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 1997;94:3290-5. [PubMed]
- Greaves MF, Maia AT, Wiemels JL, et al. Leukemia in twins: lessons in natural history. Blood 2003;102:2321-33. [PubMed]
- Zahm SH, Fraumeni JF Jr. Racial, ethnic, and gender variations in cancer risk: considerations for future epidemiologic research. Environ Health Perspect 1995;103 Suppl 8:283-6. [PubMed]
- Perera FP. Environment and cancer: who are susceptible? Science 1997;278:1068-73. [PubMed]
- Saadat M. Hardy-Weinberg equilibrium and control subjects. Kidney Int 2006;70:1374-5. [PubMed]